| Reaction Details |
|---|
 | Report a problem with these data |
| Target | N-lysine methyltransferase KMT5A |
|---|
| Ligand | BDBM50300041 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_647521 (CHEMBL1217461) |
|---|
| IC50 | >40000±n/a nM |
|---|
| Citation |  Liu, F; Chen, X; Allali-Hassani, A; Quinn, AM; Wigle, TJ; Wasney, GA; Dong, A; Senisterra, G; Chau, I; Siarheyeva, A; Norris, JL; Kireev, DB; Jadhav, A; Herold, JM; Janzen, WP; Arrowsmith, CH; Frye, SV; Brown, PJ; Simeonov, A; Vedadi, M; Jin, J Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem53:5844-57 (2010) [PubMed] Article Liu, F; Chen, X; Allali-Hassani, A; Quinn, AM; Wigle, TJ; Wasney, GA; Dong, A; Senisterra, G; Chau, I; Siarheyeva, A; Norris, JL; Kireev, DB; Jadhav, A; Herold, JM; Janzen, WP; Arrowsmith, CH; Frye, SV; Brown, PJ; Simeonov, A; Vedadi, M; Jin, J Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem53:5844-57 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| N-lysine methyltransferase KMT5A |
|---|
| Name: | N-lysine methyltransferase KMT5A |
|---|
| Synonyms: | KMT5A | KMT5A_HUMAN | N-lysine methyltransferase SETD8 | PRSET7 | SET domain-containing protein 8 (SetD8) | SET07 | SET8 | SETD8 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 42919.11 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9NQR1 |
|---|
| Residue: | 393 |
|---|
| Sequence: | MGEGGAAAALVAAAAAAAAAAAAVVAGQRRRRLGRRARCHGPGRAAGGKMSKPCAVEAAA
AAVAATAPGPEMVERRGPGRPRTDGENVFTGQSKIYSYMSPNKCSGMRFPLQEENSVTHH
EVKCQGKPLAGIYRKREEKRNAGNAVRSAMKSEEQKIKDARKGPLVPFPNQKSEAAEPPK
TPPSSCDSTNAAIAKQALKKPIKGKQAPRKKAQGKTQQNRKLTDFYPVRRSSRKSKAELQ
SEERKRIDELIESGKEEGMKIDLIDGKGRGVIATKQFSRGDFVVEYHGDLIEITDAKKRE
ALYAQDPSTGCYMYYFQYLSKTYCVDATRETNRLGRLINHSKCGNCQTKLHDIDGVPHLI
LIASRDIAAGEELLYDYGDRSKASIEAHPWLKH
|
|
|
|---|
| BDBM50300041 |
|---|
| n/a |
|---|
| Name | BDBM50300041 |
|---|
| Synonyms: | 7-(3-(dimethylamino)propoxy)-6-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinazolin-4-amine | 7-[3-(dimethylamino)propoxy]-6-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinazolin-4-amine | CHEMBL576781 | UNC0224 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H43N7O2 |
|---|
| Mol. Mass. | 485.6653 |
|---|
| SMILES | COc1cc2c(NC3CCN(C)CC3)nc(nc2cc1OCCCN(C)C)N1CCCN(C)CC1 |
|---|
| Structure | 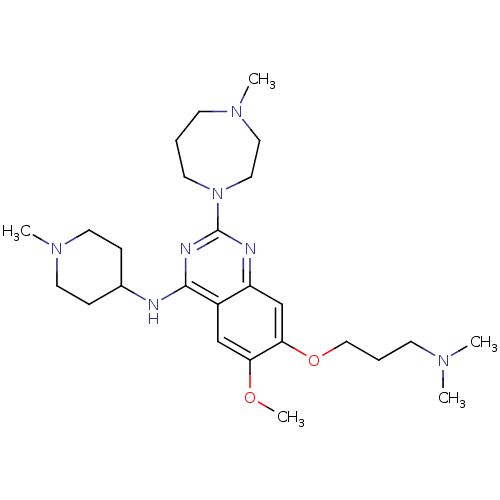
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Liu, F; Chen, X; Allali-Hassani, A; Quinn, AM; Wigle, TJ; Wasney, GA; Dong, A; Senisterra, G; Chau, I; Siarheyeva, A; Norris, JL; Kireev, DB; Jadhav, A; Herold, JM; Janzen, WP; Arrowsmith, CH; Frye, SV; Brown, PJ; Simeonov, A; Vedadi, M; Jin, J Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem53:5844-57 (2010) [PubMed] Article
Liu, F; Chen, X; Allali-Hassani, A; Quinn, AM; Wigle, TJ; Wasney, GA; Dong, A; Senisterra, G; Chau, I; Siarheyeva, A; Norris, JL; Kireev, DB; Jadhav, A; Herold, JM; Janzen, WP; Arrowsmith, CH; Frye, SV; Brown, PJ; Simeonov, A; Vedadi, M; Jin, J Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J Med Chem53:5844-57 (2010) [PubMed] Article