| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM60622 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_650759 (CHEMBL1227519) |
|---|
| IC50 | 4±n/a nM |
|---|
| Citation |  Long, JZ; Li, W; Booker, L; Burston, JJ; Kinsey, SG; Schlosburg, JE; Pavón, FJ; Serrano, AM; Selley, DE; Parsons, LH; Lichtman, AH; Cravatt, BF Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol5:37-44 (2008) [PubMed] Article Long, JZ; Li, W; Booker, L; Burston, JJ; Kinsey, SG; Schlosburg, JE; Pavón, FJ; Serrano, AM; Selley, DE; Parsons, LH; Lichtman, AH; Cravatt, BF Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol5:37-44 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM60622 |
|---|
| n/a |
|---|
| Name | BDBM60622 |
|---|
| Synonyms: | BDBM50300355 | US11753371, Compound JZL-184 | US9133148, A |
|---|
| Type | n/a |
|---|
| Emp. Form. | C27H24N2O9 |
|---|
| Mol. Mass. | 520.4875 |
|---|
| SMILES | OC(C1CCN(CC1)C(=O)Oc1ccc(cc1)[N+]([O-])=O)(c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
|---|
| Structure | 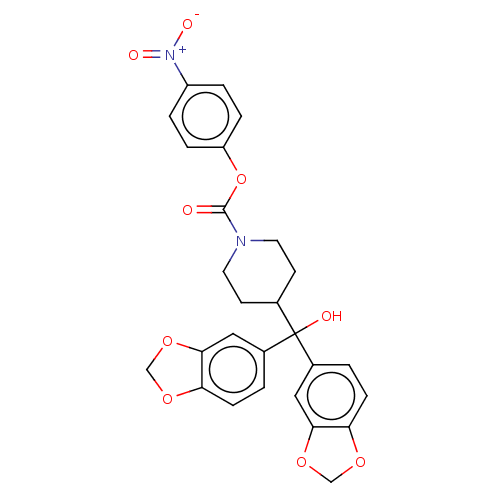
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Long, JZ; Li, W; Booker, L; Burston, JJ; Kinsey, SG; Schlosburg, JE; Pavón, FJ; Serrano, AM; Selley, DE; Parsons, LH; Lichtman, AH; Cravatt, BF Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol5:37-44 (2008) [PubMed] Article
Long, JZ; Li, W; Booker, L; Burston, JJ; Kinsey, SG; Schlosburg, JE; Pavón, FJ; Serrano, AM; Selley, DE; Parsons, LH; Lichtman, AH; Cravatt, BF Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol5:37-44 (2008) [PubMed] Article