| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prolyl hydroxylase EGLN2 |
|---|
| Ligand | BDBM26113 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_665252 (CHEMBL1260833) |
|---|
| IC50 | 624800±n/a nM |
|---|
| Citation |  Conejo-Garcia, A; McDonough, MA; Loenarz, C; McNeill, LA; Hewitson, KS; Ge, W; Liénard, BM; Schofield, CJ; Clifton, IJ Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor. Bioorg Med Chem Lett20:6125-8 (2010) [PubMed] Article Conejo-Garcia, A; McDonough, MA; Loenarz, C; McNeill, LA; Hewitson, KS; Ge, W; Liénard, BM; Schofield, CJ; Clifton, IJ Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor. Bioorg Med Chem Lett20:6125-8 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prolyl hydroxylase EGLN2 |
|---|
| Name: | Prolyl hydroxylase EGLN2 |
|---|
| Synonyms: | EGLN2 | EGLN2_HUMAN | EIT6 | Egl nine homolog 2 | Estrogen-induced tag 6 | HIF prolyl hydroxylase | HIF-PH1 | HIF-prolyl hydroxylase 1 | HPH-3 | Hypoxia-inducible factor prolyl hydroxylase | PHD1 | Prolyl hydroxylase domain-containing protein 1 | Prolyl hydroxylase domain-containing protein 1 (PHD1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 43657.01 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q96KS0 |
|---|
| Residue: | 407 |
|---|
| Sequence: | MDSPCQPQPLSQALPQLPGSSSEPLEPEPGRARMGVESYLPCPLLPSYHCPGVPSEASAG
SGTPRATATSTTASPLRDGFGGQDGGELRPLQSEGAAALVTKGCQRLAAQGARPEAPKRK
WAEDGGDAPSPSKRPWARQENQEAEREGGMSCSCSSGSGEASAGLMEEALPSAPERLALD
YIVPCMRYYGICVKDSFLGAALGGRVLAEVEALKRGGRLRDGQLVSQRAIPPRSIRGDQI
AWVEGHEPGCRSIGALMAHVDAVIRHCAGRLGSYVINGRTKAMVACYPGNGLGYVRHVDN
PHGDGRCITCIYYLNQNWDVKVHGGLLQIFPEGRPVVANIEPLFDRLLIFWSDRRNPHEV
KPAYATRYAITVWYFDAKERAAAKDKYQLASGQKGVQVPVSQPPTPT
|
|
|
|---|
| BDBM26113 |
|---|
| n/a |
|---|
| Name | BDBM26113 |
|---|
| Synonyms: | 2,4 PDCA | cid_10365 | pyridine carboxylate, 6a | pyridine-2,4-dicarboxylic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C7H5NO4 |
|---|
| Mol. Mass. | 167.1189 |
|---|
| SMILES | OC(=O)c1ccnc(c1)C(O)=O |
|---|
| Structure | 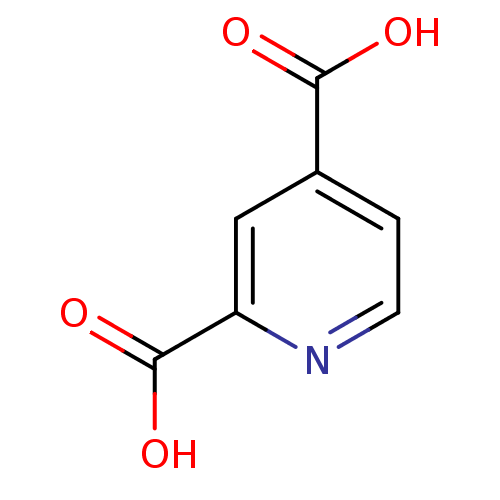
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Conejo-Garcia, A; McDonough, MA; Loenarz, C; McNeill, LA; Hewitson, KS; Ge, W; Liénard, BM; Schofield, CJ; Clifton, IJ Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor. Bioorg Med Chem Lett20:6125-8 (2010) [PubMed] Article
Conejo-Garcia, A; McDonough, MA; Loenarz, C; McNeill, LA; Hewitson, KS; Ge, W; Liénard, BM; Schofield, CJ; Clifton, IJ Structural basis for binding of cyclic 2-oxoglutarate analogues to factor-inhibiting hypoxia-inducible factor. Bioorg Med Chem Lett20:6125-8 (2010) [PubMed] Article