| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Proteinase-activated receptor 1 |
|---|
| Ligand | BDBM50261110 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_675462 (CHEMBL1273599) |
|---|
| Ki | 8±n/a nM |
|---|
| Citation |  Xia, Y; Chackalamannil, S; Greenlee, WJ; Wang, Y; Hu, Z; Root, Y; Wong, J; Kong, J; Ahn, HS; Boykow, G; Hsieh, Y; Kurowski, S; Chintala, M Discovery of a vorapaxar analog with increased aqueous solubility. Bioorg Med Chem Lett20:6676-9 (2010) [PubMed] Article Xia, Y; Chackalamannil, S; Greenlee, WJ; Wang, Y; Hu, Z; Root, Y; Wong, J; Kong, J; Ahn, HS; Boykow, G; Hsieh, Y; Kurowski, S; Chintala, M Discovery of a vorapaxar analog with increased aqueous solubility. Bioorg Med Chem Lett20:6676-9 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Proteinase-activated receptor 1 |
|---|
| Name: | Proteinase-activated receptor 1 |
|---|
| Synonyms: | CF2R | Coagulation factor II receptor | F2R | PAR-1 | PAR1 | PAR1_HUMAN | Proteinase activated receptor 1 | Proteinase-activated receptor 1 (PAR-1) | TR | Thrombin receptor | Thrombin receptor/ Proteinase-activated receptor 1(Par-1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 47450.07 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P25116 |
|---|
| Residue: | 425 |
|---|
| Sequence: | MGPRRLLLVAACFSLCGPLLSARTRARRPESKATNATLDPRSFLLRNPNDKYEPFWEDEE
KNESGLTEYRLVSINKSSPLQKQLPAFISEDASGYLTSSWLTLFVPSVYTGVFVVSLPLN
IMAIVVFILKMKVKKPAVVYMLHLATADVLFVSVLPFKISYYFSGSDWQFGSELCRFVTA
AFYCNMYASILLMTVISIDRFLAVVYPMQSLSWRTLGRASFTCLAIWALAIAGVVPLLLK
EQTIQVPGLNITTCHDVLNETLLEGYYAYYFSAFSAVFFFVPLIISTVCYVSIIRCLSSS
AVANRSKKSRALFLSAAVFCIFIICFGPTNVLLIAHYSFLSHTSTTEAAYFAYLLCVCVS
SISCCIDPLIYYYASSECQRYVYSILCCKESSDPSSYNSSGQLMASKMDTCSSNLNNSIY
KKLLT
|
|
|
|---|
| BDBM50261110 |
|---|
| n/a |
|---|
| Name | BDBM50261110 |
|---|
| Synonyms: | CHEMBL493982 | Ethyl [(3aR,4aR,8aR,9aS)-9(S)-[(E)-2-[5-(3-fluorophenyl)-2-pyridinyl]ethenyl]dodecahydro-1(R)-methyl-3-oxonaphtho[2,3-c]furan-6(R)-yl]carbamate | VORAPAXAR |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H33FN2O4 |
|---|
| Mol. Mass. | 492.5817 |
|---|
| SMILES | CCOC(=O)N[C@@H]1CC[C@@H]2[C@H](C[C@@H]3[C@@H]([C@@H](C)OC3=O)[C@H]2C=Cc2ccc(cn2)-c2cccc(F)c2)C1 |r,w:21.23| |
|---|
| Structure | 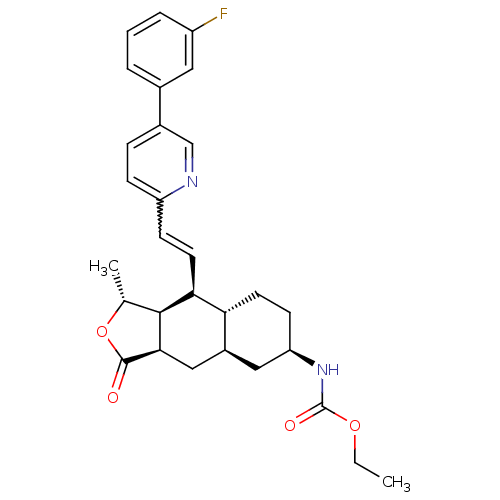
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Xia, Y; Chackalamannil, S; Greenlee, WJ; Wang, Y; Hu, Z; Root, Y; Wong, J; Kong, J; Ahn, HS; Boykow, G; Hsieh, Y; Kurowski, S; Chintala, M Discovery of a vorapaxar analog with increased aqueous solubility. Bioorg Med Chem Lett20:6676-9 (2010) [PubMed] Article
Xia, Y; Chackalamannil, S; Greenlee, WJ; Wang, Y; Hu, Z; Root, Y; Wong, J; Kong, J; Ahn, HS; Boykow, G; Hsieh, Y; Kurowski, S; Chintala, M Discovery of a vorapaxar analog with increased aqueous solubility. Bioorg Med Chem Lett20:6676-9 (2010) [PubMed] Article