

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | 5-hydroxytryptamine receptor 3A | ||
| Ligand | BDBM50329758 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_674333 (CHEMBL1274533) | ||
| Ki | 36.4±n/a nM | ||
| Citation |  Yang, Z; Fairfax, DJ; Maeng, JH; Masih, L; Usyatinsky, A; Hassler, C; Isaacson, S; Fitzpatrick, K; DeOrazio, RJ; Chen, J; Harding, JP; Isherwood, M; Dobritsa, S; Christensen, KL; Wierschke, JD; Bliss, BI; Peterson, LH; Beer, CM; Cioffi, C; Lynch, M; Rennells, WM; Richards, JJ; Rust, T; Khmelnitsky, YL; Cohen, ML; Manning, DD Discovery of 2-substituted benzoxazole carboxamides as 5-HT3 receptor antagonists. Bioorg Med Chem Lett20:6538-41 (2010) [PubMed] Article Yang, Z; Fairfax, DJ; Maeng, JH; Masih, L; Usyatinsky, A; Hassler, C; Isaacson, S; Fitzpatrick, K; DeOrazio, RJ; Chen, J; Harding, JP; Isherwood, M; Dobritsa, S; Christensen, KL; Wierschke, JD; Bliss, BI; Peterson, LH; Beer, CM; Cioffi, C; Lynch, M; Rennells, WM; Richards, JJ; Rust, T; Khmelnitsky, YL; Cohen, ML; Manning, DD Discovery of 2-substituted benzoxazole carboxamides as 5-HT3 receptor antagonists. Bioorg Med Chem Lett20:6538-41 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| 5-hydroxytryptamine receptor 3A | |||
| Name: | 5-hydroxytryptamine receptor 3A | ||
| Synonyms: | 5-HT-3 | 5-HT3 | 5-HT3A | 5-HT3R | 5-hydroxytryptamine receptor 3 (5-HT3) | 5-hydroxytryptamine receptor 3A (5-HT3a) | 5-hydroxytryptamine receptor 3A (5HT3A) | 5HT3A_HUMAN | 5HT3R | HTR3 | HTR3A | Serotonin 3 (5-HT3) receptor | Serotonin 3 receptor (5HT3) | Serotonin receptor 3A | Serotonin-gated ion channel receptor | Serotonin-gated ion channel receptor 3 | ||
| Type: | Protein | ||
| Mol. Mass.: | 55283.27 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P46098 | ||
| Residue: | 478 | ||
| Sequence: |
| ||
| BDBM50329758 | |||
| n/a | |||
| Name | BDBM50329758 | ||
| Synonyms: | (S)-2-(methylamino)-N-(quinuclidin-3-yl)benzo[d]oxazole-4-carboxamide | CHEMBL1272177 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C16H20N4O2 | ||
| Mol. Mass. | 300.3556 | ||
| SMILES | CNc1nc2c(cccc2o1)C(=O)N[C@@H]1CN2CCC1CC2 |r,wD:14.15,TLB:13:14:18.17:20.21,(2.65,-3.98,;1.89,-5.31,;.35,-5.32,;-.56,-4.06,;-2.04,-4.54,;-3.37,-3.78,;-4.7,-4.55,;-4.7,-6.09,;-3.37,-6.87,;-2.04,-6.09,;-.56,-6.57,;-3.38,-2.24,;-4.71,-1.47,;-2.04,-1.47,;-2.05,.07,;-2.24,1.45,;-.78,2.1,;.58,1.49,;.85,.09,;-.52,.73,;-.26,2.63,;-.71,3.74,)| | ||
| Structure | 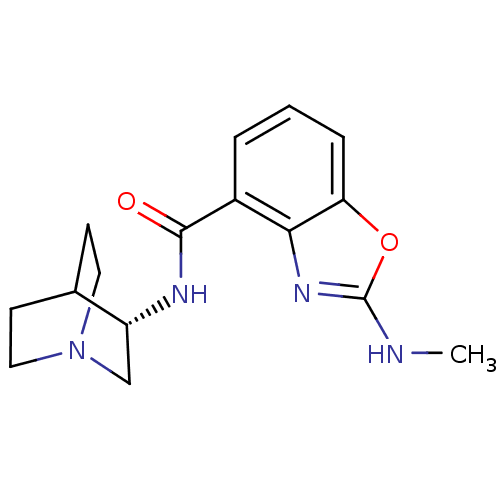
| ||