

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M2 | ||
| Ligand | BDBM50344270 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_748301 (CHEMBL1781311) | ||
| IC50 | 1.8±n/a nM | ||
| Citation |  Prat, M; Buil, MA; Fernández, MD; Castro, J; Monleón, JM; Tort, L; Casals, G; Ferrer, M; Huerta, JM; Espinosa, S; López, M; Segarra, V; Gavaldà, A; Miralpeix, M; Ramos, I; Vilella, D; González, M; Córdoba, M; Cárdenas, A; Antón, F; Beleta, J; Ryder, H Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinyl carbamates as potent and long acting muscarinic antagonists. Bioorg Med Chem Lett21:3457-61 (2011) [PubMed] Article Prat, M; Buil, MA; Fernández, MD; Castro, J; Monleón, JM; Tort, L; Casals, G; Ferrer, M; Huerta, JM; Espinosa, S; López, M; Segarra, V; Gavaldà, A; Miralpeix, M; Ramos, I; Vilella, D; González, M; Córdoba, M; Cárdenas, A; Antón, F; Beleta, J; Ryder, H Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinyl carbamates as potent and long acting muscarinic antagonists. Bioorg Med Chem Lett21:3457-61 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M2 | |||
| Name: | Muscarinic acetylcholine receptor M2 | ||
| Synonyms: | ACM2_HUMAN | CHRM2 | Cholinergic, muscarinic M2 | Muscarinic acetylcholine receptor M2 and M4 | Muscarinic acetylcholine receptor M2 and M5 | RecName: Full=Muscarinic acetylcholine receptor M2 | ||
| Type: | GPCR | ||
| Mol. Mass.: | 51730.61 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P08172 | ||
| Residue: | 466 | ||
| Sequence: |
| ||
| BDBM50344270 | |||
| n/a | |||
| Name | BDBM50344270 | ||
| Synonyms: | (R)-quinuclidin-3-yl phenyl(thiophen-2-ylmethyl)carbamate | CHEMBL1779033 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H22N2O2S | ||
| Mol. Mass. | 342.455 | ||
| SMILES | O=C(O[C@H]1CN2CCC1CC2)N(Cc1cccs1)c1ccccc1 |r,wD:3.2,(-3.52,-8.18,;-3.52,-9.72,;-2.19,-10.49,;-.85,-9.72,;.48,-10.5,;1.81,-9.74,;1.82,-8.2,;.48,-7.42,;-.86,-8.19,;-.1,-9.51,;1.03,-8.39,;-4.85,-10.49,;-4.85,-12.03,;-6.19,-12.8,;-7.6,-12.18,;-8.63,-13.33,;-7.86,-14.66,;-6.36,-14.34,;-6.19,-9.72,;-6.19,-8.18,;-7.52,-7.41,;-8.85,-8.18,;-8.85,-9.73,;-7.52,-10.49,)| | ||
| Structure | 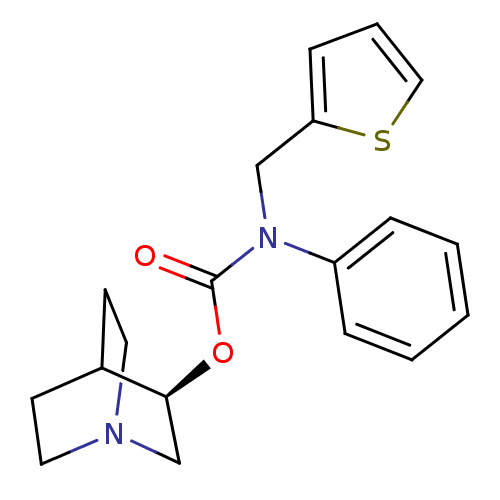
| ||