

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Corticotropin-releasing factor receptor 1 | ||
| Ligand | BDBM50272695 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_759193 (CHEMBL1811339) | ||
| Ki | 0.2±n/a nM | ||
| Citation |  Hodgetts, KJ; Ge, P; Yoon, T; De Lombaert, S; Brodbeck, R; Gulianello, M; Kieltyka, A; Horvath, RF; Kehne, JH; Krause, JE; Maynard, GD; Hoffman, D; Lee, Y; Fung, L; Doller, D Discovery of N-(1-ethylpropyl)-[3-methoxy-5-(2-methoxy-4-trifluoromethoxyphenyl)-6-methyl-pyrazin-2-yl]amine 59 (NGD 98-2): an orally active corticotropin releasing factor-1 (CRF-1) receptor antagonist. J Med Chem54:4187-206 (2011) [PubMed] Article Hodgetts, KJ; Ge, P; Yoon, T; De Lombaert, S; Brodbeck, R; Gulianello, M; Kieltyka, A; Horvath, RF; Kehne, JH; Krause, JE; Maynard, GD; Hoffman, D; Lee, Y; Fung, L; Doller, D Discovery of N-(1-ethylpropyl)-[3-methoxy-5-(2-methoxy-4-trifluoromethoxyphenyl)-6-methyl-pyrazin-2-yl]amine 59 (NGD 98-2): an orally active corticotropin releasing factor-1 (CRF-1) receptor antagonist. J Med Chem54:4187-206 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Corticotropin-releasing factor receptor 1 | |||
| Name: | Corticotropin-releasing factor receptor 1 | ||
| Synonyms: | CRF-R | CRF-R2 Alpha | CRF1 | CRFR | CRFR1 | CRFR1_HUMAN | CRH-R 1 | CRHR | CRHR1 | Corticotropin releasing factor receptor 1 | Corticotropin-releasing factor receptor 1 (CRF-1) | Corticotropin-releasing factor receptor 1 (CRF1) | Corticotropin-releasing hormone receptor 1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 50744.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34998 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50272695 | |||
| n/a | |||
| Name | BDBM50272695 | ||
| Synonyms: | 3-(4-chloro-2-morpholinothiazol-5-yl)-2,6-dimethyl-8-(pentan-3-yl)imidazo[1,2-b]pyridazine | CHEMBL498311 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H26ClN5OS | ||
| Mol. Mass. | 419.971 | ||
| SMILES | CCC(CC)c1cc(C)nn2c(c(C)nc12)-c1sc(nc1Cl)N1CCOCC1 |(8.98,-11.26,;8.98,-12.8,;7.65,-13.58,;6.32,-12.81,;4.99,-13.59,;7.66,-15.12,;6.34,-15.89,;6.33,-17.43,;5,-18.2,;7.67,-18.2,;9.01,-17.43,;10.49,-17.9,;11.4,-16.64,;12.94,-16.63,;10.48,-15.39,;9,-15.88,;10.98,-19.36,;12.45,-19.83,;12.46,-21.37,;11,-21.85,;10.09,-20.61,;8.55,-20.62,;13.71,-22.26,;13.55,-23.79,;14.79,-24.69,;16.2,-24.06,;16.36,-22.53,;15.11,-21.62,)| | ||
| Structure | 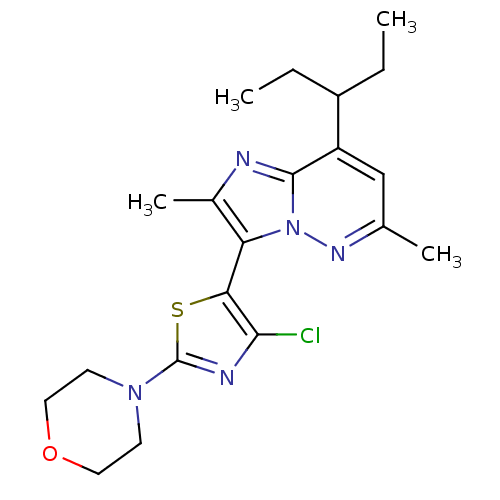
| ||