| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM50349811 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_760320 (CHEMBL1815135) |
|---|
| EC50 | 0.1±n/a nM |
|---|
| Citation |  Raval, P; Jain, M; Goswami, A; Basu, S; Gite, A; Godha, A; Pingali, H; Raval, S; Giri, S; Suthar, D; Shah, M; Patel, P Revisiting glitazars: thiophene substituted oxazole containinga-ethoxy phenylpropanoic acid derivatives as highly potent PPARa/¿ dual agonists devoid of adverse effects in rodents. Bioorg Med Chem Lett21:3103-9 (2011) [PubMed] Article Raval, P; Jain, M; Goswami, A; Basu, S; Gite, A; Godha, A; Pingali, H; Raval, S; Giri, S; Suthar, D; Shah, M; Patel, P Revisiting glitazars: thiophene substituted oxazole containinga-ethoxy phenylpropanoic acid derivatives as highly potent PPARa/¿ dual agonists devoid of adverse effects in rodents. Bioorg Med Chem Lett21:3103-9 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM50349811 |
|---|
| n/a |
|---|
| Name | BDBM50349811 |
|---|
| Synonyms: | CHEMBL1813008 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H23NO5S |
|---|
| Mol. Mass. | 401.476 |
|---|
| SMILES | CCO[C@@H](Cc1ccc(OCCc2nc(oc2C)-c2cccs2)cc1)C(O)=O |r| |
|---|
| Structure | 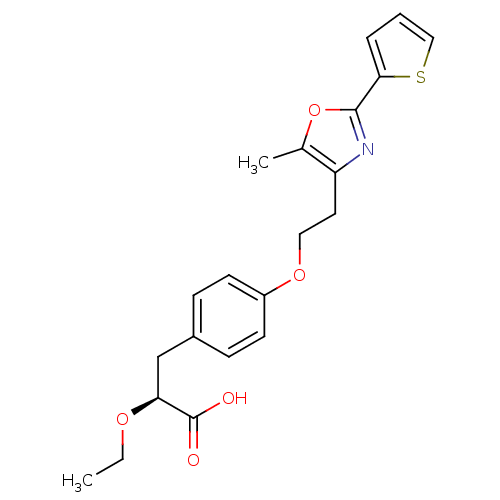
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Raval, P; Jain, M; Goswami, A; Basu, S; Gite, A; Godha, A; Pingali, H; Raval, S; Giri, S; Suthar, D; Shah, M; Patel, P Revisiting glitazars: thiophene substituted oxazole containinga-ethoxy phenylpropanoic acid derivatives as highly potent PPARa/¿ dual agonists devoid of adverse effects in rodents. Bioorg Med Chem Lett21:3103-9 (2011) [PubMed] Article
Raval, P; Jain, M; Goswami, A; Basu, S; Gite, A; Godha, A; Pingali, H; Raval, S; Giri, S; Suthar, D; Shah, M; Patel, P Revisiting glitazars: thiophene substituted oxazole containinga-ethoxy phenylpropanoic acid derivatives as highly potent PPARa/¿ dual agonists devoid of adverse effects in rodents. Bioorg Med Chem Lett21:3103-9 (2011) [PubMed] Article