

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50350728 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_762993 (CHEMBL1820810) | ||
| IC50 | 2400±n/a nM | ||
| Citation |  Bengtsson, C; Blaho, S; Saitton, DB; Brickmann, K; Broddefalk, J; Davidsson, O; Drmota, T; Folmer, R; Hallberg, K; Hallén, S; Hovland, R; Isin, E; Johannesson, P; Kull, B; Larsson, LO; Löfgren, L; Nilsson, KE; Noeske, T; Oakes, N; Plowright, AT; Schnecke, V; Ståhlberg, P; Sörme, P; Wan, H; Wellner, E; Oster, L Design of small molecule inhibitors of acetyl-CoA carboxylase 1 and 2 showing reduction of hepatic malonyl-CoA levels in vivo in obese Zucker rats. Bioorg Med Chem19:3039-53 (2011) [PubMed] Article Bengtsson, C; Blaho, S; Saitton, DB; Brickmann, K; Broddefalk, J; Davidsson, O; Drmota, T; Folmer, R; Hallberg, K; Hallén, S; Hovland, R; Isin, E; Johannesson, P; Kull, B; Larsson, LO; Löfgren, L; Nilsson, KE; Noeske, T; Oakes, N; Plowright, AT; Schnecke, V; Ståhlberg, P; Sörme, P; Wan, H; Wellner, E; Oster, L Design of small molecule inhibitors of acetyl-CoA carboxylase 1 and 2 showing reduction of hepatic malonyl-CoA levels in vivo in obese Zucker rats. Bioorg Med Chem19:3039-53 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50350728 | |||
| n/a | |||
| Name | BDBM50350728 | ||
| Synonyms: | CHEMBL1818291 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H38N4O3 | ||
| Mol. Mass. | 502.6477 | ||
| SMILES | CC(C)(C)OC(=O)NC[C@H]1CC[C@H](CNC(=O)c2cc(nc3ccccc23)-c2cccc(CN)c2)CC1 |r,wU:9.8,wD:12.12,(-10.3,5.18,;-8.97,5.96,;-8.98,7.5,;-10.31,6.73,;-7.64,5.19,;-6.3,5.97,;-6.31,7.51,;-4.97,5.2,;-4.96,3.66,;-3.63,2.89,;-2.3,3.67,;-.97,2.89,;-.97,1.35,;.37,.58,;.36,-.96,;1.7,-1.73,;3.03,-.96,;1.69,-3.27,;3.02,-4.03,;3.03,-5.57,;1.69,-6.35,;.36,-5.58,;-.97,-6.33,;-2.29,-5.57,;-2.29,-4.03,;-.96,-3.27,;.36,-4.04,;4.36,-6.34,;4.36,-7.88,;5.69,-8.65,;7.03,-7.87,;7.02,-6.33,;8.35,-5.55,;9.69,-6.31,;5.68,-5.56,;-2.3,.59,;-3.63,1.35,)| | ||
| Structure | 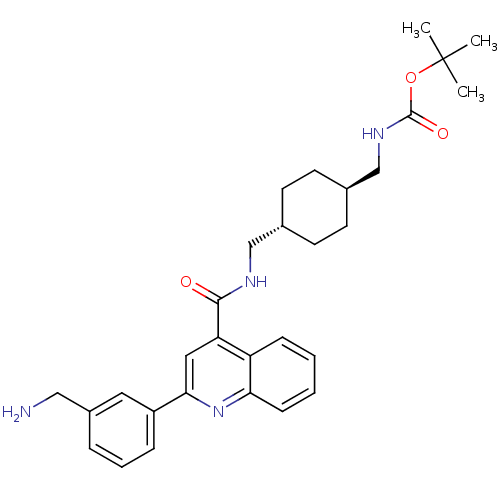
| ||