| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Insulin receptor |
|---|
| Ligand | BDBM50306682 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_767354 (CHEMBL1825466) |
|---|
| IC50 | 102±n/a nM |
|---|
| Citation |  Cui, JJ; Tran-Dubé, M; Shen, H; Nambu, M; Kung, PP; Pairish, M; Jia, L; Meng, J; Funk, L; Botrous, I; McTigue, M; Grodsky, N; Ryan, K; Padrique, E; Alton, G; Timofeevski, S; Yamazaki, S; Li, Q; Zou, H; Christensen, J; Mroczkowski, B; Bender, S; Kania, RS; Edwards, MP Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem54:6342-63 (2011) [PubMed] Article Cui, JJ; Tran-Dubé, M; Shen, H; Nambu, M; Kung, PP; Pairish, M; Jia, L; Meng, J; Funk, L; Botrous, I; McTigue, M; Grodsky, N; Ryan, K; Padrique, E; Alton, G; Timofeevski, S; Yamazaki, S; Li, Q; Zou, H; Christensen, J; Mroczkowski, B; Bender, S; Kania, RS; Edwards, MP Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem54:6342-63 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Insulin receptor |
|---|
| Name: | Insulin receptor |
|---|
| Synonyms: | INSR | INSR protein | INSR_HUMAN | Insulin receptor (IR) | Insulin receptor beta subunit |
|---|
| Type: | Receptor Tyrosine Kinase |
|---|
| Mol. Mass.: | 156322.60 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P06213 |
|---|
| Residue: | 1382 |
|---|
| Sequence: | MATGGRRGAAAAPLLVAVAALLLGAAGHLYPGEVCPGMDIRNNLTRLHELENCSVIEGHL
QILLMFKTRPEDFRDLSFPKLIMITDYLLLFRVYGLESLKDLFPNLTVIRGSRLFFNYAL
VIFEMVHLKELGLYNLMNITRGSVRIEKNNELCYLATIDWSRILDSVEDNYIVLNKDDNE
ECGDICPGTAKGKTNCPATVINGQFVERCWTHSHCQKVCPTICKSHGCTAEGLCCHSECL
GNCSQPDDPTKCVACRNFYLDGRCVETCPPPYYHFQDWRCVNFSFCQDLHHKCKNSRRQG
CHQYVIHNNKCIPECPSGYTMNSSNLLCTPCLGPCPKVCHLLEGEKTIDSVTSAQELRGC
TVINGSLIINIRGGNNLAAELEANLGLIEEISGYLKIRRSYALVSLSFFRKLRLIRGETL
EIGNYSFYALDNQNLRQLWDWSKHNLTITQGKLFFHYNPKLCLSEIHKMEEVSGTKGRQE
RNDIALKTNGDQASCENELLKFSYIRTSFDKILLRWEPYWPPDFRDLLGFMLFYKEAPYQ
NVTEFDGQDACGSNSWTVVDIDPPLRSNDPKSQNHPGWLMRGLKPWTQYAIFVKTLVTFS
DERRTYGAKSDIIYVQTDATNPSVPLDPISVSNSSSQIILKWKPPSDPNGNITHYLVFWE
RQAEDSELFELDYCLKGLKLPSRTWSPPFESEDSQKHNQSEYEDSAGECCSCPKTDSQIL
KELEESSFRKTFEDYLHNVVFVPRKTSSGTGAEDPRPSRKRRSLGDVGNVTVAVPTVAAF
PNTSSTSVPTSPEEHRPFEKVVNKESLVISGLRHFTGYRIELQACNQDTPEERCSVAAYV
SARTMPEAKADDIVGPVTHEIFENNVVHLMWQEPKEPNGLIVLYEVSYRRYGDEELHLCV
SRKHFALERGCRLRGLSPGNYSVRIRATSLAGNGSWTEPTYFYVTDYLDVPSNIAKIIIG
PLIFVFLFSVVIGSIYLFLRKRQPDGPLGPLYASSNPEYLSASDVFPCSVYVPDEWEVSR
EKITLLRELGQGSFGMVYEGNARDIIKGEAETRVAVKTVNESASLRERIEFLNEASVMKG
FTCHHVVRLLGVVSKGQPTLVVMELMAHGDLKSYLRSLRPEAENNPGRPPPTLQEMIQMA
AEIADGMAYLNAKKFVHRDLAARNCMVAHDFTVKIGDFGMTRDIYETDYYRKGGKGLLPV
RWMAPESLKDGVFTTSSDMWSFGVVLWEITSLAEQPYQGLSNEQVLKFVMDGGYLDQPDN
CPERVTDLMRMCWQFNPKMRPTFLEIVNLLKDDLHPSFPEVSFFHSEENKAPESEELEME
FEDMENVPLDRSSHCQREEAGGRDGGSSLGFKRSYEEHIPYTHMNGGKKNGRILTLPRSN
PS
|
|
|
|---|
| BDBM50306682 |
|---|
| n/a |
|---|
| Name | BDBM50306682 |
|---|
| Synonyms: | (R)-3-(1-(2,6-dichloro-3-fluorophenyl)ethoxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine | 3-(2,6-dichloro-3-fluorobenzyloxy)-5-(1-(piperidin-4-yl)-1H-pyrazol-4-yl)pyridin-2-amine | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine | CHEMBL601719 | CRIZOTINIB | PF-2341066 | US10370379, Crizotinib | US10543199, Compound Crizotinib | US10780082, Compound Crizotinib | US11059827, Compound Crizotinib | US11517561, Compound Crizotinib | US9126941, PF-2341066 | US9199944, Crizotinib | US9226923, Crizotinib |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H22Cl2FN5O |
|---|
| Mol. Mass. | 450.337 |
|---|
| SMILES | C[C@@H](Oc1cc(cnc1N)-c1cnn(c1)C1CCNCC1)c1c(Cl)ccc(F)c1Cl |r| |
|---|
| Structure | 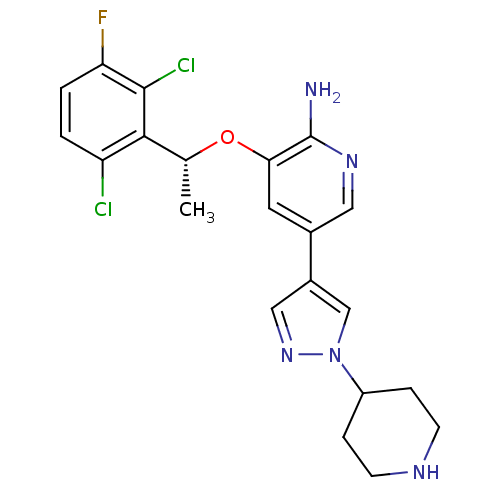
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cui, JJ; Tran-Dubé, M; Shen, H; Nambu, M; Kung, PP; Pairish, M; Jia, L; Meng, J; Funk, L; Botrous, I; McTigue, M; Grodsky, N; Ryan, K; Padrique, E; Alton, G; Timofeevski, S; Yamazaki, S; Li, Q; Zou, H; Christensen, J; Mroczkowski, B; Bender, S; Kania, RS; Edwards, MP Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem54:6342-63 (2011) [PubMed] Article
Cui, JJ; Tran-Dubé, M; Shen, H; Nambu, M; Kung, PP; Pairish, M; Jia, L; Meng, J; Funk, L; Botrous, I; McTigue, M; Grodsky, N; Ryan, K; Padrique, E; Alton, G; Timofeevski, S; Yamazaki, S; Li, Q; Zou, H; Christensen, J; Mroczkowski, B; Bender, S; Kania, RS; Edwards, MP Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem54:6342-63 (2011) [PubMed] Article