

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Diacylglycerol O-acyltransferase 1 | ||
| Ligand | BDBM50355146 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_772695 (CHEMBL1837163) | ||
| IC50 | 98±n/a nM | ||
| Citation |  Dow, RL; Andrews, M; Aspnes, GE; Balan, G; Michael Gibbs, E; Guzman-Perez, A; Karki, K; Laperle, JL; Li, JC; Litchfield, J; Munchhof, MJ; Perreault, C; Patel, L Design and synthesis of potent, orally-active DGAT-1 inhibitors containing a dioxino[2,3-d]pyrimidine core. Bioorg Med Chem Lett21:6122-5 (2011) [PubMed] Article Dow, RL; Andrews, M; Aspnes, GE; Balan, G; Michael Gibbs, E; Guzman-Perez, A; Karki, K; Laperle, JL; Li, JC; Litchfield, J; Munchhof, MJ; Perreault, C; Patel, L Design and synthesis of potent, orally-active DGAT-1 inhibitors containing a dioxino[2,3-d]pyrimidine core. Bioorg Med Chem Lett21:6122-5 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Diacylglycerol O-acyltransferase 1 | |||
| Name: | Diacylglycerol O-acyltransferase 1 | ||
| Synonyms: | ACAT-related gene product 1 | AGRP1 | Acyl coA-diacylglycerol acyl transferase 1 (DGAT1) | DGAT | DGAT1 | DGAT1_HUMAN | Diacylglycerol Acyltransferase (DGAT1) | Diacylglycerol O-acyltransferase 1 | Diacylglycerol O-acyltransferase 1 (DGAT1) | Diglyceride acyltransferase | ||
| Type: | Protein | ||
| Mol. Mass.: | 55297.82 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O75907 | ||
| Residue: | 488 | ||
| Sequence: |
| ||
| BDBM50355146 | |||
| n/a | |||
| Name | BDBM50355146 | ||
| Synonyms: | CHEMBL1834555 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H24N4O3 | ||
| Mol. Mass. | 368.4296 | ||
| SMILES | NC(=O)C[C@H]1CC[C@@H](CC1)c1ccc(cc1)[C@H]1COc2ncnc(N)c2O1 |r,wU:7.10,16.17,wD:4.3,(42.86,-8.87,;42.9,-7.33,;44.23,-6.55,;41.56,-6.57,;40.23,-7.35,;38.89,-6.6,;37.56,-7.38,;37.58,-8.92,;38.93,-9.67,;40.25,-8.89,;36.26,-9.7,;36.28,-11.24,;34.96,-12.02,;33.62,-11.27,;33.59,-9.73,;34.91,-8.94,;32.29,-12.03,;32.29,-13.58,;30.95,-14.36,;29.61,-13.6,;28.27,-14.36,;26.94,-13.59,;26.94,-12.05,;28.27,-11.28,;28.26,-9.74,;29.6,-12.04,;30.94,-11.25,)| | ||
| Structure | 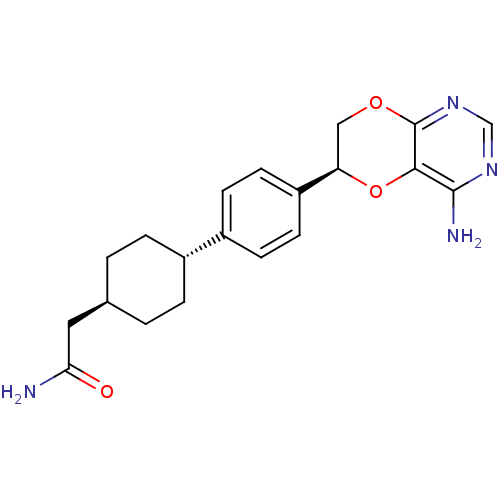
| ||