| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Ribosomal protein S6 kinase beta-1 |
|---|
| Ligand | BDBM13336 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_774386 (CHEMBL1908603) |
|---|
| Kd | >10000±n/a nM |
|---|
| Citation |  Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Ribosomal protein S6 kinase beta-1 |
|---|
| Name: | Ribosomal protein S6 kinase beta-1 |
|---|
| Synonyms: | 70 kDa ribosomal protein S6 kinase 1 | 70 kDa ribosomal protein S6 kinase 1 (P70s6k) | 70 kDa ribosomal protein kinase (p70S6K) | KS6B1_HUMAN | Protein kinase 70S6K (T412E) | RAC-alpha serine/threonine-protein kinase/Ribosomal protein S6 kinase beta-1 | RPS6KB1 | Ribosomal protein S6 kinase (P70S6K) | Ribosomal protein S6 kinase I | S6K1 | STK14A | p70 S6 ribosomal protein kinase 1 | p70S6K1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 59139.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | His-tagged S6K1. |
|---|
| Residue: | 525 |
|---|
| Sequence: | MRRRRRRDGFYPAPDFRDREAEDMAGVFDIDLDQPEDAGSEDELEEGGQLNESMDHGGVG
PYELGMEHCEKFEISETSVNRGPEKIRPECFELLRVLGKGGYGKVFQVRKVTGANTGKIF
AMKVLKKAMIVRNAKDTAHTKAERNILEEVKHPFIVDLIYAFQTGGKLYLILEYLSGGEL
FMQLEREGIFMEDTACFYLAEISMALGHLHQKGIIYRDLKPENIMLNHQGHVKLTDFGLC
KESIHDGTVTHTFCGTIEYMAPEILMRSGHNRAVDWWSLGALMYDMLTGAPPFTGENRKK
TIDKILKCKLNLPPYLTQEARDLLKKLLKRNAASRLGAGPGDAGEVQAHPFFRHINWEEL
LARKVEPPFKPLLQSEEDVSQFDSKFTRQTPVDSPDDSTLSESANQVFLGFTYVAPSVLE
SVKEKFSFEPKIRSPRRFIGSPRTPVSPVKFSPGDFWGRGASASTANPQTPVEYPMETSG
IEQMDVTMSGEASAPLPIRQPNSGPYKKQAFPMISKRPEHLRMNL
|
|
|
|---|
| BDBM13336 |
|---|
| n/a |
|---|
| Name | BDBM13336 |
|---|
| Synonyms: | 4-[4-(4-fluorophenyl)-2-(4-methanesulfinylphenyl)-1H-imidazol-5-yl]pyridine | 4-[4-(4-fluorophenyl)-2-(4-methylsulfinylphenyl)-1H-imidazol-5-yl]pyridine | 4-{4-(4-fluorophenyl)-2-[4-(methylsulfinyl)phenyl]-1H-imidazol-5-yl}pyridine | CHEMBL10 | SB-203580 | SB203580 | cid_176155 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H16FN3OS |
|---|
| Mol. Mass. | 377.435 |
|---|
| SMILES | CS(=O)c1ccc(cc1)-c1nc(c([nH]1)-c1ccc(F)cc1)-c1ccncc1 |
|---|
| Structure | 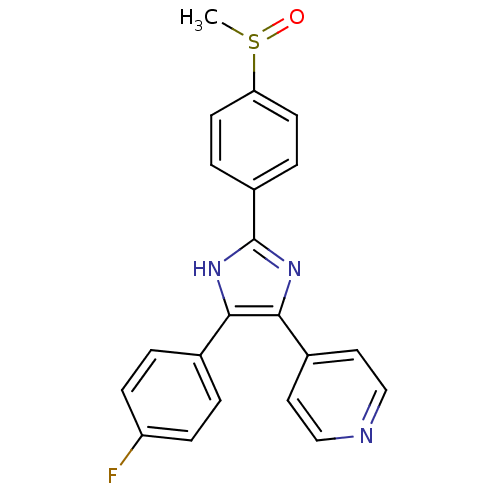
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article
Davis, MI; Hunt, JP; Herrgard, S; Ciceri, P; Wodicka, LM; Pallares, G; Hocker, M; Treiber, DK; Zarrinkar, PP Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol29:1046-51 (2011) [PubMed] Article