| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase Lck |
|---|
| Ligand | BDBM13216 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_776844 (CHEMBL1913941) |
|---|
| IC50 | <1±n/a nM |
|---|
| Citation |  Andreani, A; Granaiola, M; Leoni, A; Locatelli, A; Morigi, R; Rambaldi, M; Varoli, L; Lannigan, D; Smith, J; Scudiero, D; Kondapaka, S; Shoemaker, RH Imidazo[2,1-b]thiazole guanylhydrazones as RSK2 inhibitors. Eur J Med Chem46:4311-23 (2011) [PubMed] Article Andreani, A; Granaiola, M; Leoni, A; Locatelli, A; Morigi, R; Rambaldi, M; Varoli, L; Lannigan, D; Smith, J; Scudiero, D; Kondapaka, S; Shoemaker, RH Imidazo[2,1-b]thiazole guanylhydrazones as RSK2 inhibitors. Eur J Med Chem46:4311-23 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein kinase Lck |
|---|
| Name: | Tyrosine-protein kinase Lck |
|---|
| Synonyms: | 2.7.10.2 | LCK | LCK_HUMAN | LSK | Leukocyte C-terminal Src kinase | Lymphocyte cell-specific protein-tyrosine kinase | Lymphocyte-specific protein tyrosine kinase | P56-LCK | Protein YT16 | Proto-oncogene Lck | Proto-oncogene tyrosine-protein kinase LCK | Src/Lck kinase | T cell-specific protein-tyrosine kinase |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 57987.83 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P06239 |
|---|
| Residue: | 509 |
|---|
| Sequence: | MGCGCSSHPEDDWMENIDVCENCHYPIVPLDGKGTLLIRNGSEVRDPLVTYEGSNPPASP
LQDNLVIALHSYEPSHDGDLGFEKGEQLRILEQSGEWWKAQSLTTGQEGFIPFNFVAKAN
SLEPEPWFFKNLSRKDAERQLLAPGNTHGSFLIRESESTAGSFSLSVRDFDQNQGEVVKH
YKIRNLDNGGFYISPRITFPGLHELVRHYTNASDGLCTRLSRPCQTQKPQKPWWEDEWEV
PRETLKLVERLGAGQFGEVWMGYYNGHTKVAVKSLKQGSMSPDAFLAEANLMKQLQHQRL
VRLYAVVTQEPIYIITEYMENGSLVDFLKTPSGIKLTINKLLDMAAQIAEGMAFIEERNY
IHRDLRAANILVSDTLSCKIADFGLARLIEDNEYTAREGAKFPIKWTAPEAINYGTFTIK
SDVWSFGILLTEIVTHGRIPYPGMTNPEVIQNLERGYRMVRPDNCPEELYQLMRLCWKER
PEDRPTFDYLRSVLEDFFTATEGQYQPQP
|
|
|
|---|
| BDBM13216 |
|---|
| n/a |
|---|
| Name | BDBM13216 |
|---|
| Synonyms: | BMS-354825 | CHEMBL1421 | DASATINIB | N-(2-Chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide | N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide | US10294227, Code Dasatinib | US20230348453, Compound A8 | cid_3062316 | med.21724, Compound Dasatinib |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H26ClN7O2S |
|---|
| Mol. Mass. | 488.006 |
|---|
| SMILES | Cc1nc(Nc2ncc(s2)C(=O)Nc2c(C)cccc2Cl)cc(n1)N1CCN(CCO)CC1 |
|---|
| Structure | 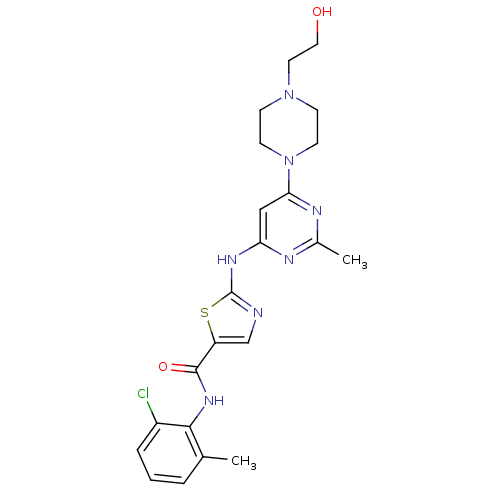
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Andreani, A; Granaiola, M; Leoni, A; Locatelli, A; Morigi, R; Rambaldi, M; Varoli, L; Lannigan, D; Smith, J; Scudiero, D; Kondapaka, S; Shoemaker, RH Imidazo[2,1-b]thiazole guanylhydrazones as RSK2 inhibitors. Eur J Med Chem46:4311-23 (2011) [PubMed] Article
Andreani, A; Granaiola, M; Leoni, A; Locatelli, A; Morigi, R; Rambaldi, M; Varoli, L; Lannigan, D; Smith, J; Scudiero, D; Kondapaka, S; Shoemaker, RH Imidazo[2,1-b]thiazole guanylhydrazones as RSK2 inhibitors. Eur J Med Chem46:4311-23 (2011) [PubMed] Article