

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Dipeptidyl peptidase 4 | ||
| Ligand | BDBM50356587 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_776111 (CHEMBL1912807) | ||
| Ki | 0.46±n/a nM | ||
| Citation |  Wang, W; Devasthale, P; Wang, A; Harrity, T; Egan, D; Morgan, N; Cap, M; Fura, A; Klei, HE; Kish, K; Weigelt, C; Sun, L; Levesque, P; Li, YX; Zahler, R; Kirby, MS; Hamann, LG 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site. Bioorg Med Chem Lett21:6646-51 (2011) [PubMed] Article Wang, W; Devasthale, P; Wang, A; Harrity, T; Egan, D; Morgan, N; Cap, M; Fura, A; Klei, HE; Kish, K; Weigelt, C; Sun, L; Levesque, P; Li, YX; Zahler, R; Kirby, MS; Hamann, LG 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site. Bioorg Med Chem Lett21:6646-51 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Dipeptidyl peptidase 4 | |||
| Name: | Dipeptidyl peptidase 4 | ||
| Synonyms: | ADABP | ADCP2 | Adenosine deaminase complexing protein 2 | CD26 | CD_antigen=CD26 | DPP IV | DPP4 | DPP4_HUMAN | DPPIV | Dipeptidyl peptidase 4 (DDP-IV) | Dipeptidyl peptidase 4 (DPP IV) | Dipeptidyl peptidase 4 (DPP-4) | Dipeptidyl peptidase 4 (DPP4) | Dipeptidyl peptidase 4 (DPPIV) | Dipeptidyl peptidase 4 membrane form | Dipeptidyl peptidase 4 soluble form | Dipeptidyl peptidase IV | Dipeptidyl peptidase IV (DDP-4) | Dipeptidyl peptidase IV (DDP-IV) | Dipeptidyl peptidase IV (DPP IV) | Dipeptidyl peptidase IV membrane form | Dipeptidyl peptidase IV soluble form | Dipeptidyl peptidase-IV (DPP-4) | Dipeptidyl peptidase-IV (DPP-IV) | T-cell activation antigen CD26 | TP103 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 88271.01 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P27487 | ||
| Residue: | 766 | ||
| Sequence: |
| ||
| BDBM50356587 | |||
| n/a | |||
| Name | BDBM50356587 | ||
| Synonyms: | CHEMBL1910121 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H25Cl2N5O4S | ||
| Mol. Mass. | 526.436 | ||
| SMILES | Cc1nc2C(=O)N(CC(=O)N3CCN(CC3)S(C)(=O)=O)Cc2c(c1CN)-c1ccc(Cl)cc1Cl |(48.61,-29.07,;47.28,-29.84,;45.95,-29.08,;44.62,-29.86,;43.16,-29.38,;42.68,-27.91,;42.25,-30.62,;40.72,-30.62,;39.95,-31.96,;40.72,-33.29,;38.41,-31.96,;37.64,-30.63,;36.11,-30.63,;35.33,-31.96,;36.1,-33.29,;37.65,-33.3,;33.79,-31.95,;33.03,-30.62,;32.45,-32.72,;33.78,-33.49,;43.15,-31.87,;44.62,-31.4,;45.95,-32.17,;47.29,-31.39,;48.62,-32.16,;49.96,-31.39,;45.95,-33.7,;44.62,-34.47,;44.62,-36.01,;45.95,-36.78,;45.95,-38.32,;47.29,-36,;47.28,-34.47,;48.61,-33.69,)| | ||
| Structure | 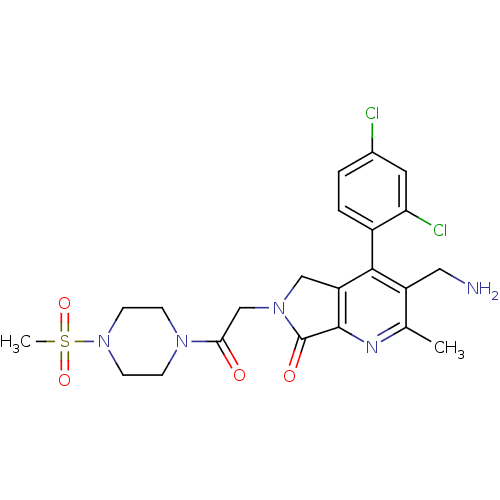
| ||