| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Laccase |
|---|
| Ligand | BDBM26985 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_786157 (CHEMBL1919991) |
|---|
| IC50 | 279000±n/a nM |
|---|
| Citation |  Dalisay, DS; Saludes, JP; Molinski, TF Ptilomycalin A inhibits laccase and melanization in Cryptococcus neoformans. Bioorg Med Chem19:6654-7 (2011) [PubMed] Article Dalisay, DS; Saludes, JP; Molinski, TF Ptilomycalin A inhibits laccase and melanization in Cryptococcus neoformans. Bioorg Med Chem19:6654-7 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Laccase |
|---|
| Name: | Laccase |
|---|
| Synonyms: | n/a |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 55472.53 |
|---|
| Organism: | Trametes versicolor |
|---|
| Description: | ChEMBL_786157 |
|---|
| Residue: | 520 |
|---|
| Sequence: | MSRFHSLLAFVVASLAAVAHAGIGPVADLTITNAAVSPDGFSRQAVVVNGGTPGPLITGN
MGDRFQLNVIDNLTDHTMLKSTSIHWHGFFQKGTNWADGPAFINQCPISSGHSFLYDFQV
PDQAGTFWYHSHLSTQYCDGLRGPFVVYDPNDPAADLYDVDNDDTVITLADWYHVAAKLG
PAFPLGADATLINGKGRSPSTTTADLTVISVTPGKRYRFRLVSLSCDPNHTFSIDGHNMT
IIETDSINTAPLVVDSIQIFAAQRYSFVLEANQAVDNYWIRANPSFGNVGFTGGINSAIL
RYDGAAAIEPTTTQTTSTEPLNEVNLHPLVATAVPGSPAAGGVDLAINMAFNFNGTNFFI
NGASFTPPTVPVLLQIISGAQNAQDLLPSGSVYSLPSNADIEISFPATAAAPGAPHPFHL
HGHAFAVVRSAGSTVYNYDNPIFRDVVSTGTPAAGDNVTIRFRTDNPGPWFLHCHIDFHL
EAGFAVVFAEDIPDVASANPVPQAWSDLCPTYDARDPSDQ
|
|
|
|---|
| BDBM26985 |
|---|
| n/a |
|---|
| Name | BDBM26985 |
|---|
| Synonyms: | 2-triaza-1,2-diyne | CHEMBL79455 | CHEMBL89295 | N3(-1) | azide |
|---|
| Type | Ion |
|---|
| Emp. Form. | N3 |
|---|
| Mol. Mass. | 42.0206 |
|---|
| SMILES | [N-]=[N+]=[N-] |
|---|
| Structure | 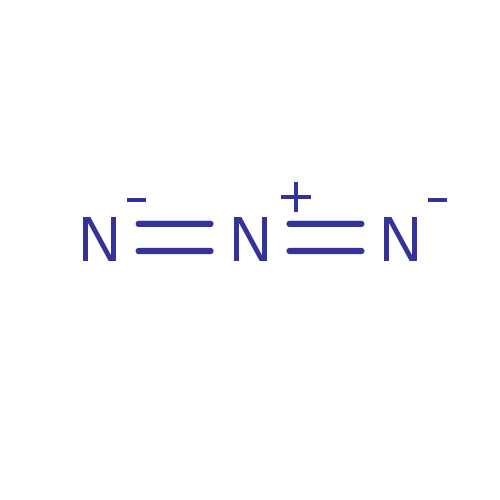
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Dalisay, DS; Saludes, JP; Molinski, TF Ptilomycalin A inhibits laccase and melanization in Cryptococcus neoformans. Bioorg Med Chem19:6654-7 (2011) [PubMed] Article
Dalisay, DS; Saludes, JP; Molinski, TF Ptilomycalin A inhibits laccase and melanization in Cryptococcus neoformans. Bioorg Med Chem19:6654-7 (2011) [PubMed] Article