

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Metabotropic glutamate receptor 4 | ||
| Ligand | BDBM55094 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_788858 (CHEMBL1924928) | ||
| EC50 | 491±n/a nM | ||
| Citation |  Jones, CK; Engers, DW; Thompson, AD; Field, JR; Blobaum, AL; Lindsley, SR; Zhou, Y; Gogliotti, RD; Jadhav, S; Zamorano, R; Bogenpohl, J; Smith, Y; Morrison, R; Daniels, JS; Weaver, CD; Conn, PJ; Lindsley, CW; Niswender, CM; Hopkins, CR Discovery, synthesis, and structure-activity relationship development of a series of N-4-(2,5-dioxopyrrolidin-1-yl)phenylpicolinamides (VU0400195, ML182): characterization of a novel positive allosteric modulator of the metabotropic glutamate receptor 4 (mGlu(4)) with oral efficacy in an antiparkin J Med Chem54:7639-47 (2011) [PubMed] Article Jones, CK; Engers, DW; Thompson, AD; Field, JR; Blobaum, AL; Lindsley, SR; Zhou, Y; Gogliotti, RD; Jadhav, S; Zamorano, R; Bogenpohl, J; Smith, Y; Morrison, R; Daniels, JS; Weaver, CD; Conn, PJ; Lindsley, CW; Niswender, CM; Hopkins, CR Discovery, synthesis, and structure-activity relationship development of a series of N-4-(2,5-dioxopyrrolidin-1-yl)phenylpicolinamides (VU0400195, ML182): characterization of a novel positive allosteric modulator of the metabotropic glutamate receptor 4 (mGlu(4)) with oral efficacy in an antiparkin J Med Chem54:7639-47 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Metabotropic glutamate receptor 4 | |||
| Name: | Metabotropic glutamate receptor 4 | ||
| Synonyms: | GRM4_RAT | Gprc1d | Grm4 | Metabotropic glutamate receptor | Metabotropic glutamate receptor 4 (mGlu4) | Mglur4 | mGlu4 | metabotropic glutamate 4 | metabotropic glutamate 4a | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 101849.79 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | P31423 | ||
| Residue: | 912 | ||
| Sequence: |
| ||
| BDBM55094 | |||
| n/a | |||
| Name | BDBM55094 | ||
| Synonyms: | N-[3-chloro-4-(2,5-diketo-3-phenyl-pyrrolidino)phenyl]picolinamide | N-[3-chloro-4-(2,5-dioxo-3-phenyl-1-pyrrolidinyl)phenyl]-2-pyridinecarboxamide | N-[3-chloro-4-(2,5-dioxo-3-phenylpyrrolidin-1-yl)phenyl]pyridine-2-carboxamide | N-[4-[2,5-bis(oxidanylidene)-3-phenyl-pyrrolidin-1-yl]-3-chloranyl-phenyl]pyridine-2-carboxamide | VU0400071-3 | cid_45110765 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H16ClN3O3 | ||
| Mol. Mass. | 405.834 | ||
| SMILES | Oc1cc(c(O)n1-c1ccc(NC(=O)c2ccccn2)cc1Cl)-c1ccccc1 |(3.08,3.07,;4.54,3.55,;5.02,5.01,;6.56,5.01,;7.04,3.55,;8.5,3.07,;5.79,2.64,;5.79,1.1,;7.12,.33,;7.12,-1.21,;5.79,-1.98,;5.79,-3.52,;7.12,-4.29,;8.46,-3.52,;7.12,-5.83,;8.46,-6.6,;8.46,-8.14,;7.12,-8.91,;5.79,-8.14,;5.79,-6.6,;4.46,-1.21,;4.46,.33,;3.12,1.1,;7.47,6.26,;9,6.1,;9.9,7.34,;9.28,8.75,;7.74,8.91,;6.84,7.66,)| | ||
| Structure | 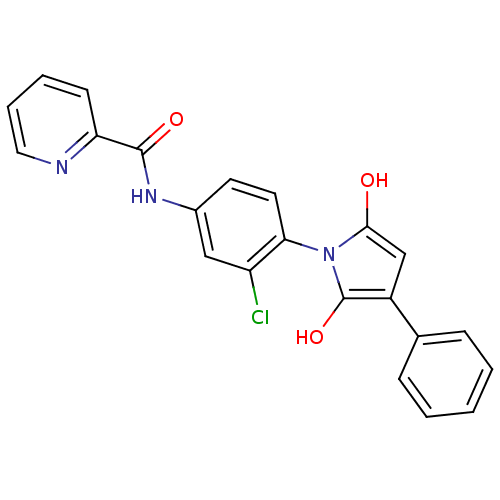
| ||