

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Substance-P receptor | ||
| Ligand | BDBM50070554 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_142731 (CHEMBL745035) | ||
| IC50 | 1.9±n/a nM | ||
| Citation |  Kubota, H; Okamoto, Y; Fujii, M; Ikeda, K; Takeuchi, M; Shibanuma, T; Isomura, Y Synthesis and NK1 receptor antagonistic activity of (+/-)-1-acyl-3-(3,4- dichlorophenyl)-3-[2-(spiro-substituted piperidin-1'-yl)ethyl]piperidines. Bioorg Med Chem Lett8:1541-6 (1999) [PubMed] Kubota, H; Okamoto, Y; Fujii, M; Ikeda, K; Takeuchi, M; Shibanuma, T; Isomura, Y Synthesis and NK1 receptor antagonistic activity of (+/-)-1-acyl-3-(3,4- dichlorophenyl)-3-[2-(spiro-substituted piperidin-1'-yl)ethyl]piperidines. Bioorg Med Chem Lett8:1541-6 (1999) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Substance-P receptor | |||
| Name: | Substance-P receptor | ||
| Synonyms: | NK1R_CAVPO | Neurokinin 1 receptor | Neurokinin NK1 | TAC1R | TACR1 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 46261.42 | ||
| Organism: | GUINEA PIG | ||
| Description: | Neurokinin NK1 TACR1 GUINEA PIG::P30547 | ||
| Residue: | 407 | ||
| Sequence: |
| ||
| BDBM50070554 | |||
| n/a | |||
| Name | BDBM50070554 | ||
| Synonyms: | 1'-(2-{3-(3,4-dichlorophenyl)-1-[2-(3-isopropoxyphenyl)acetyl]hexahydro-3-pyridinyl}ethyl)-1'-methylspiro[1,2,3,4-tetrahydroisoquinoline-1,4'-(hexahydropyridine)ium]-3-one iodide | CHEMBL288123 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C38H46Cl2N3O3 | ||
| Mol. Mass. | 663.696 | ||
| SMILES | CC(C)Oc1cccc(CC(=O)N2CCCC(CC[N+]3(C)CCC4(CC3)NC(=O)Cc3ccccc43)(C2)c2ccc(Cl)c(Cl)c2)c1 |(14.47,-5.41,;13.13,-4.64,;13.15,-3.1,;11.78,-5.38,;10.46,-4.61,;10.48,-3.07,;9.14,-2.28,;7.79,-3.03,;7.79,-4.58,;6.44,-5.35,;5.12,-4.57,;3.77,-5.32,;5.12,-3.02,;6.48,-2.26,;6.48,-.72,;5.16,.06,;3.83,-.71,;2.49,-1.49,;1.16,-.74,;-.18,-1.52,;.23,-3.02,;-1.5,-2.03,;-2.93,-1.52,;-3.83,-2.78,;-2.66,-2.39,;-.92,-2.93,;-4.6,-4.1,;-6.13,-4.1,;-6.9,-5.41,;-6.88,-2.78,;-6.11,-1.46,;-6.87,-.13,;-6.11,1.19,;-4.59,1.18,;-3.82,-.14,;-4.59,-1.46,;3.81,-2.25,;3.81,.84,;2.48,1.6,;2.46,3.14,;3.79,3.92,;3.77,5.49,;5.12,3.17,;6.44,3.96,;5.15,1.63,;9.11,-5.37,)| | ||
| Structure | 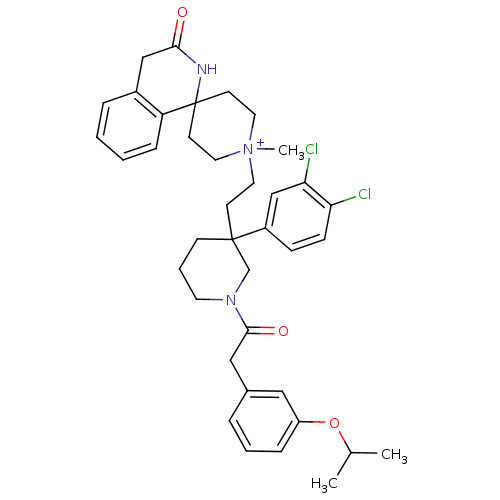
| ||