

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Adenosine receptor A2a | ||
| Ligand | BDBM50085674 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_31973 | ||
| Ki | 0.050000±n/a nM | ||
| Citation |  Keeling, SE; Albinson, FD; Ayres, BE; Butchers, PR; Chambers, CL; Cherry, PC; Ellis, F; Ewan, GB; Gregson, M; Knight, J; Mills, K; Ravenscroft, P; Reynolds, LH; Sanjar, S; Sheehan, MJ The discovery and synthesis of highly potent, A2a receptor agonists. Bioorg Med Chem Lett10:403-6 (2000) [PubMed] Keeling, SE; Albinson, FD; Ayres, BE; Butchers, PR; Chambers, CL; Cherry, PC; Ellis, F; Ewan, GB; Gregson, M; Knight, J; Mills, K; Ravenscroft, P; Reynolds, LH; Sanjar, S; Sheehan, MJ The discovery and synthesis of highly potent, A2a receptor agonists. Bioorg Med Chem Lett10:403-6 (2000) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Adenosine receptor A2a | |||
| Name: | Adenosine receptor A2a | ||
| Synonyms: | A2A adenosine receptor (hA2A) | AA2AR_HUMAN | ADENOSINE A2 | ADENOSINE A2a | ADORA2 | ADORA2A | Adenosine A2A receptor (A2AAR) | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 44716.46 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P29274 | ||
| Residue: | 412 | ||
| Sequence: |
| ||
| BDBM50085674 | |||
| n/a | |||
| Name | BDBM50085674 | ||
| Synonyms: | (2S,3S,4R,5R)-5-[2-(4-Amino-cyclohexylamino)-6-(2,2-diphenyl-ethylamino)-purin-9-yl]-3,4-dihydroxy-tetrahydro-furan-2-carboxylic acid ethylamide; TFA | CHEMBL337080 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C32H40N8O4 | ||
| Mol. Mass. | 600.7112 | ||
| SMILES | CCNC(=O)[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(NCC(c3ccccc3)c3ccccc3)nc(N[C@H]3CC[C@H](N)CC3)nc12 |wU:7.12,5.4,35.37,wD:8.8,10.11,38.41,(-2.14,-10.42,;-1.05,-9.34,;-1.45,-7.85,;-.35,-6.77,;-.7,-5.26,;1.12,-7.24,;2.36,-6.34,;3.6,-7.24,;3.12,-8.72,;4.04,-9.95,;1.59,-8.72,;.69,-9.95,;3.38,-4.67,;4.3,-3.42,;3.38,-2.18,;1.92,-2.66,;.59,-1.89,;.57,-.35,;1.92,.42,;1.89,1.96,;3.23,2.73,;4.55,1.96,;5.89,2.73,;5.89,4.28,;4.53,5.03,;3.21,4.26,;.57,2.71,;.57,4.26,;-.77,5.01,;-2.1,4.24,;-2.09,2.7,;-.77,1.93,;-.75,-2.66,;-.75,-4.22,;-2.09,-4.98,;-2.87,-3.64,;-4.41,-3.66,;-5.2,-2.31,;-4.42,-.95,;-5.2,.37,;-2.87,-.95,;-2.09,-2.29,;.59,-4.98,;1.92,-4.2,)| | ||
| Structure | 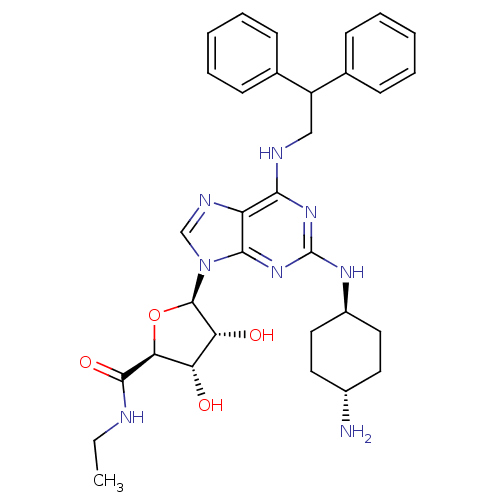
| ||