| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 1/2/3/4/5/6/7/8/9/11/Polyamine deacetylase HDAC10 |
|---|
| Ligand | BDBM50366962 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_87707 (CHEMBL877727) |
|---|
| IC50 | 84±n/a nM |
|---|
| Citation |  Nishino, N; Yoshikawa, D; Watanabe, LA; Kato, T; Jose, B; Komatsu, Y; Sumida, Y; Yoshida, M Synthesis and histone deacetylase inhibitory activity of cyclic tetrapeptides containing a retrohydroxamate as zinc ligand. Bioorg Med Chem Lett14:2427-31 (2004) [PubMed] Article Nishino, N; Yoshikawa, D; Watanabe, LA; Kato, T; Jose, B; Komatsu, Y; Sumida, Y; Yoshida, M Synthesis and histone deacetylase inhibitory activity of cyclic tetrapeptides containing a retrohydroxamate as zinc ligand. Bioorg Med Chem Lett14:2427-31 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 1/2/3/4/5/6/7/8/9/11/Polyamine deacetylase HDAC10 |
|---|
| Name: | Histone deacetylase 1/2/3/4/5/6/7/8/9/11/Polyamine deacetylase HDAC10 |
|---|
| Synonyms: | Histone deacetylase |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 87707 |
|---|
| Components: | This complex has 11 components. |
|---|
| Component 1 |
| Name: | Histone deacetylase 2 |
|---|
| Synonyms: | 3.5.1.98 | HD2 | HDAC2_MOUSE | Hdac2 | Histone deacetylase 2 | YY1 transcription factor-binding protein | Yy1bp |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 55294.47 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_117015 |
|---|
| Residue: | 488 |
|---|
| Sequence: | MAYSQGGGKKKVCYYYDGDIGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKA
TAEEMTKYHSDEYIKFLRSIRPDNMSEYSKQMQRFNVGEDCPVFDGLFEFCQLSTGGSVA
GAVKLNRQQTDMAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHH
GDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNFPMRDGIDDESYGQ
IFKPIISKVMEMYQPSAVVLQCGADSLSGDRLGCFNLTVKGHAKCVEVAKTFNLPLLMLG
GGGYTIRNVARCWTYETAVALDCEIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTPEYM
EKIKQRLFENLRMLPHAPGVQMQAIPEDAVHEDSGDEDGEDPDKRISIRASDKRIACDEE
FSDSEDEGEGGRRNVADHKKGAKKARIEEDKKETEDKKTDVKEEDKSKDNSGEKTDPKGA
KSEQLSNP
|
|
|
|---|
| Component 2 |
| Name: | Histone deacetylase 1 |
|---|
| Synonyms: | HD1 | HDAC1_MOUSE | Hdac1 | Histone deacetylase 1/2 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55062.26 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | O09106 |
|---|
| Residue: | 482 |
|---|
| Sequence: | MAQTQGTKRKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKAN
AEEMTKYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGEDCPVFDGLFEFCQLSTGGSVAS
AVKLNKQQTDIAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHG
DGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAI
FKPVMSKVMEMFQPSAVVLQCGSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNLPMLMLGG
GGYTIRNVARCWTYETAVALDTEIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTNEYLE
KIKQRLFENLRMLPHAPGVQMQAIPEDAIPEESGDEDEEDPDKRISICSSDKRIACEEEF
SDSDEEGEGGRKNSSNFKKAKRVKTEDEKEKDPEEKKEVTEEEKTKEEKPEAKGVKEEVK
LA
|
|
|
|---|
| Component 3 |
| Name: | Histone deacetylase 6 |
|---|
| Synonyms: | HD6 | HDAC6_MOUSE | Hdac6 | Histone Deacetylase 6 (HDAC6) | Histone deacetylase | Histone deacetylase mHDA2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 125762.28 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_1460074 |
|---|
| Residue: | 1149 |
|---|
| Sequence: | MTSTGQDSSTRQRKSRHNPQSPLQESSATLKRGGKKCAVPHSSPNLAEVKKKGKMKKLSQ
PAEEDLVVGLQGLDLNPETRVPVGTGLVFDEQLNDFHCLWDDSFPESPERLHAIREQLIL
EGLLGRCVSFQARFAEKEELMLVHSLEYIDLMETTQYMNEGELRVLAETYDSVYLHPNSY
SCACLATGSVLRLVDALMGAEIRNGMAVIRPPGHHAQHNLMDGYCMFNHLAVAARYAQKK
HRIQRVLIVDWDVHHGQGTQFIFDQDPSVLYFSIHRYEHGRFWPHLKASNWSTIGFGQGQ
GYTINVPWNQTGMRDADYIAAFLHILLPVASEFQPQLVLVAAGFDALHGDPKGEMAATPA
GFAHLTHLLMGLAGGKLILSLEGGYNLRALAKGVSASLHTLLGDPCPMLESCVVPCASAQ
TSIYCTLEALEPFWEVLERSVETQEEDEVEEAVLEEEEEEGGWEATALPMDTWPLLQNRT
GLVYDEKMMSHCNLWDNHHPETPQRILRIMCHLEEVGLAARCLILPARPALDSELLTCHS
AEYVEHLRTTEKMKTRDLHREGANFDSIYICPSTFACAKLATGAACRLVEAVLSGEVLNG
IAVVRPPGHHAEPNAACGFCFFNSVAVAARHAQIIAGRALRILIVDWDVHHGNGTQHIFE
DDPSVLYVSLHRYDRGTFFPMGDEGASSQVGRDAGIGFTVNVPWNGPRMGDADYLAAWHR
LVLPIAYEFNPELVLISAGFDAAQGDPLGGCQVTPEGYAHLTHLLMGLAGGRIILILEGG
YNLASISESMAACTHSLLGDPPPQLTLLRPPQSGALVSISEVIQVHRKYWRSLRLMKMED
KEECSSSRLVIKKLPPTASPVSAKEMTTPKGKVPEESVRKTIAALPGKESTLGQAKSKMA
KAVLAQGQSSEQAAKGTTLDLATSKETVGGATTDLWASAAAPENFPNQTTSVEALGETEP
TPPASHTNKQTTGASPLQGVTAQQSLQLGVLSTLELSREAEEAHDSEEGLLGEAAGGQDM
NSLMLTQGFGDFNTQDVFYAVTPLSWCPHLMAVCPIPAAGLDVSQPCKTCGTVQENWVCL
TCYQVYCSRYVNAHMVCHHEASEHPLVLSCVDLSTWCYVCQAYVHHEDLQDVKNAAHQNK
FGEDMPHSH
|
|
|
|---|
| Component 4 |
| Name: | Histone deacetylase 5 |
|---|
| Synonyms: | HD5 | HDAC5_MOUSE | Hdac5 | Histone deacetylase | Histone deacetylase mHDA1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 120932.97 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_87889 |
|---|
| Residue: | 1113 |
|---|
| Sequence: | MNSPNESDGMSGREPSLGILPRTPLHSIPVAVEVKPVLPGAMPSSMGGGGGGSPSPVELR
GALAGPMDPALREQQLQQELLVLKQQQQLQKQLLFAEFQKQHDHLTRQHEVQLQKHLKQQ
QEMLAAKRQQELEQQRQREQQRQEELEKQRLEQQLLILRNKEKSKESAIASTEVKLRLQE
FLLSKSKEPTPGGLNHSLPQHPKCWGAHHASLDQSSPPQSGPPGTPPSYKLPLLGPYDSR
DDFPLRKTASEPNLKVRSRLKQKVAERRSSPLLRRKDGTVISTFKKRAVEITGTGPGVSS
VCNSAPGSGPSSPNSSHSTIAENGFTGSVPNIPTEMIPQHRALPLDSSPNQFSLYTSPSL
PNISLGLQATVTVTNSHLTASPKLSTQQEAERQALQSLRQGGTLTGKFMSTSSIPGCLLG
VALEGDTSPHGHASLLQHVCSWTGRQQSTLIAVPLHGQSPLVTGERVATSMRTVGKLPRH
RPLSRTQSSPLPQSPQALQQLVMQQQHQQFLEKQKQQQMQLGKILTKTGELSRQPTTHPE
ETEEELTEQQEALLGEGALTIPREGSTESESTQEDLEEEEEEEEEEEEDCIQVKDEDGES
GPDEGPDLEESSAGYKKLFADAQQLQPLQVYQAPLSLATVPHQALGRTQSSPAAPGSMKS
PTDQPTVVKHLFTTGVVYDTFMLKHQCMCGNTHVHPEHAGRIQSIWSRLQETGLLGKCER
IRGRKATLDEIQTVHSEYHTLLYGTSPLNRQKLDSKKLLGPISQKMYAMLPCGGIGVDSD
TVWNEMHSSSAVRMAVGCLVELAFKVAAGELKNGFAIIRPPGHHAEESTAMGFCFFNSVA
ITAKLLQQKLSVGKVLIVDWDIHHGNGTQQAFYNDPSVLYISLHRYDNGNFFPGSGAPEE
VGGGPGVGYNVNVAWTGGVDPPIGDVEYLTAFRTVVMPIAQEFSPDVVLVSAGFDAVEGH
LSPLGGYSVTARCFGHLTRQLMTLAGGRVVLALEGGHDLTAICDASEACVSALLSVELQP
LDEAVLQQKPSVNAVATLEKVIEIQSKHWSCVQRFAAGLGCSLREAQTGEKEEAETVSAM
ALLSVGAEQAQAVATQEHSPRPAEEPMEQEPAL
|
|
|
|---|
| Component 5 |
| Name: | Histone deacetylase 4 |
|---|
| Synonyms: | 3.5.1.98 | HD4 | HDAC4_MOUSE | Hdac4 | Histone deacetylase 4 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 118571.33 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_116900 |
|---|
| Residue: | 1076 |
|---|
| Sequence: | MSSQSHPDGLSGRDQPVELLNPARVNHMPSTVDVATALPLQVAPTAVPMDLRLDHQFSLP
LEPALREQQLQQELLALKQKQQIQRQILIAEFQRQHEQLSRQHEAQLHEHIKQQQEMLAM
KHQQELLEHQRKLERHRQEQELEKQHREQKLQQLKNKEKGKESAVASTEVKMKLQEFVLN
KKKALAHRNLNHCISSDPRYWYGKTQHSSLDQSSPPQSGVSASYNHPVLGMYDAKDDFPL
RKTASEPNLKLRSRLKQKVAERRSSPLLRRKDGPVATALKKRPLDVTDSACSSAPGSGPS
SPNSSSGNVSTENGIAPTVPSAPAETSLAHRLVTREGSVAPLPLYTSPSLPNITLGLPAT
GPAAGAAGQQDAERLALPALQQRILFPGTHLTPYLSTSPLERDGAAAHNPLLQHMVLLEQ
PPTQTPLVTGLGALPLHSQSLVGADRVSPSIHKLRQHRPLGRTQSAPLPQNAQALQHLVI
QQQHQQFLEKHKQQFQQQQLHLSKIISKPSEPPRQPESHPEETEEELREHQALLDEPYLD
RLPGQKEPSLAGVQVKQEPIESEEEEAEATRETEPGQRPATEQELLFRQQALLLEQQRIH
QLRNYQASMEAAGIPVSFGSHRPLSRAQSSPASATFPMSVQEPPTKPRFTTGLVYDTLML
KHQCTCGNTNSHPEHAGRIQSIWSRLQETGLRGKCECIRGRKATLEELQTVHSEAHTLLY
GTNPLNRQKLDSSLTSVFVRLPCGGVGVDSDTIWNEVHSSGAARLAVGCVVELVFKVATG
ELKNGFAVVRPPGHHAEESTPMGFCYFNSVAVAAKLLQQRLNVSKILIVDWDVHHGNGTQ
QAFYNDPNVLYMSLHRYDDGNFFPGSGAPDEVGTGPGVGFNVNMAFTGGLEPPMGDAEYL
AAFRTVVMPIANEFAPDVVLVSSGFDAVEGHPTPLGGYNLSAKCFGYLTKQLMGLAGGRL
VLALEGGHDLTAICDASEACVSALLGNELEPLPEKVLHQRPNANAVHSMEKVMDIHSKYW
RCLQRLSSTVGHSLIEAQKCEKEEAETVTAMASLSVGVKPAEKRSEEEPMEEEPPL
|
|
|
|---|
| Component 6 |
| Name: | Histone deacetylase 9 |
|---|
| Synonyms: | 3.5.1.98 | HD7b | HD9 | HDAC9_MOUSE | Hdac7b | Hdac9 | Hdrp | Histone deacetylase 7B | Histone deacetylase 9 | Histone deacetylase-related protein | MEF2-interacting transcription repressor MITR | Mitr |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 65701.37 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_116900 |
|---|
| Residue: | 588 |
|---|
| Sequence: | MHSMISSVDVKSEVPMGLEPISPLDLRTDLRMMMPVVDPVVREKQLQQELLLIQQQQQIQ
KQLLIAEFQKQHENLTRQHQAQLQEHIKELLAIKQQQELLEKEQKLEQQRQEQEVERHRR
EQQLPPLRGKDRGRERAVASTEVKQKLQEFLLSKSATKDTPTNGKNHSVGRHPKLWYTAA
HHTSLDQSSPPLSGTSPSYKYTLPGAQDSKDDFPLRKTASEPNLKVRSRLKQKVAERRSS
PLLRRKDGNLVTSFKKRVFEVAESSVSSSSPGSGPSSPNNGPAGNVTENEASALPPTPHP
EQLVPQQRILIHEDSMNLLSLYTSPSLPNITLGLPAVPSPLNASNSLKDKQKCETQMLRQ
GVPLPSQYGSSIAASSSHVHVAMEGKPNSSHQALLQHLLLKEQMRQQKLLVAGGVPLHPQ
SPLATKERISPGIRGTHKLPRHRPLNRTQSAPLPQSTLAQLVIQQQHQQFLEKQKQYQQQ
IHMNKLLSKSIEQLKQPGSHLEEAEEELQGDQSMEDRAASKDNSARSDSSACVEDTLGQV
GAVKVKEEPVDSDEDAQIQEMECGEQAAFMQQVIGKDLAPGFVIKVII
|
|
|
|---|
| Component 7 |
| Name: | Histone deacetylase 7 |
|---|
| Synonyms: | 3.5.1.98 | HD7 | HD7a | HDAC7_MOUSE | Hdac7 | Hdac7a | Histone deacetylase 7 | Histone deacetylase 7A |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 101302.81 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_116900 |
|---|
| Residue: | 938 |
|---|
| Sequence: | MHSPGAGCPALQPDTPGSQPQPMDLRVGQRPTVEPPPEPALLTLQHPQRLHRHLFLAGLH
QQQRSAEPMRLSMDPPMPELQGGQQEQELRQLLNKDKSKRSAVASSVVKQKLAEVILKKQ
QAALERTVHPSSPSIPYRTLEPLDTEGAARSVLSSFLPPVPSLPTEPPEHFPLRKTVSEP
NLKLRYKPKKSLERRKNPLLRKESAPPSLRRRPAETLGDSSPSSSSTPASGCSSPNDSEH
GPNPALGSEADGDRRTHSTLGPRGPVLGNPHAPLFLHHGLEPEAGGTLPSRLQPILLLDP
SVSHAPLWTVPGLGPLPFHFAQPLLTTERLSGSGLHRPLNRTRSEPLPPSATASPLLAPL
QPRQDRLKPHVQLIKPAISPPQRPAKPSEKPRLRQIPSAEDLETDGGGVGPMANDGLEHR
ESGRGPPEGRGSISLQQHQQVPPWEQQHLAGRLSQGSPGDSVLIPLAQVGHRPLSRTQSS
PAAPVSLLSPEPTCQTQVLNSSETPATGLVYDSVMLKHQCSCGDNSKHPEHAGRIQSIWS
RLQERGLRSQCECLRGRKASLEELQSVHSERHVLLYGTNPLSRLKLDNGKLTGLLAQRTF
VMLPCGGVGVDTDTIWNELHSSNAARWAAGSVTDLAFKVASRELKNGFAVVRPPGHHADH
STAMGFCFFNSVAIACRQLQQHGKASKILIVDWDVHHGNGTQQTFYQDPSVLYISLHRHD
DGNFFPGSGAVDEVGTGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRIVVMPIAREFAPDL
VLVSAGFDAAEGHPAPLGGYHVSAKCFGYMTQQLMNLAGGAVVLALEGGHDLTAICDASE
ACVAALLGNKVDPLSEESWKQKPNLSAIRSLEAVVRVHRKYWGCMQRLASCPDSWLPRVP
GADAEVEAVTALASLSVGILAEDRPSERLVEEEEPMNL
|
|
|
|---|
| Component 8 |
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | 3.5.1.98 | HD3 | HDAC3_MOUSE | Hdac3 | Histone deacetylase 3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 48348.05 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_116900 |
|---|
| Residue: | 424 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
SQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKES
|
|
|
|---|
| Component 9 |
| Name: | Histone deacetylase 8 |
|---|
| Synonyms: | HDAC8_MOUSE | Hdac8 | Histone deacetylase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 41767.71 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_88030 |
|---|
| Residue: | 377 |
|---|
| Sequence: | MEMPEEPANSGHSLPPVYIYSPEYVSICDSLVKVPKRASMVHSLIEAYALHKQMRIVKPK
VASMEEMATFHTDAYLQHLQKVSQEGDEDHPDSIEYGLGYDCPATEGIFDYAAAIGGGTI
TAAQCLIDGKCKVAINWSGGWHHAKKDEASGFCYLNDAVLGILRLRRKFDRILYVDLDLH
HGDGVEDAFSFTSKVMTVSLHKFSPGFFPGTGDMSDVGLGKGRYYSVNVPIQDGIQDEKY
YHICESVLKEVYQAFNPKAVVLQLGADTIAGDPMCSFNMTPVGIGKCLKYVLQWQLATLI
LGGGGYNLANTARCWTYLTGVILGKTLSSEIPDHEFFTAYGPDYVLEITPSCRPDRNEPH
RIQQILNYIKGNLKHVV
|
|
|
|---|
| Component 10 |
| Name: | Histone deacetylase 11 |
|---|
| Synonyms: | 3.5.1.98 | HD11 | HDA11_MOUSE | Hdac11 | Histone deacetylase 11 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 39160.64 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_116900 |
|---|
| Residue: | 347 |
|---|
| Sequence: | MPHATQLYQHVPEKRWPIVYSPRYNITFMGLEKLHPFDAGKWGKVINFLKEEKLLSDGML
VEAREASEEDLLVVHTRRYLNELKWSFVVATITEIPPVIFLPNFLVQRKVLRPLRTQTGG
TIMAGKLAVERGWAINVGGGFHHCSSDRGGGFCAYADITLAIKFLFERVEGISRATIIDL
DAHQGNGHERDFMGDKRVYIMDVYNRHIYPGDRFAKEAIRRKVELEWGTEDEEYLEKVER
NVRRSLQEHLPDVVVYNAGTDVLEGDRLGGLSISPAGIVKRDEVVFRVVRAHDIPILMVT
SGGYQKRTARIIADSILNLHDLGLIGPEFPCVSAQNSGIPLLSCAVP
|
|
|
|---|
| Component 11 |
| Name: | Polyamine deacetylase HDAC10 |
|---|
| Synonyms: | 3.5.1.98 | HD10 | HDA10_MOUSE | Hdac10 | Histone deacetylase 10 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 72091.98 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_116900 |
|---|
| Residue: | 666 |
|---|
| Sequence: | MGTALVYHEDMTATRLLWDDPECEIECPERLTAALDGLRQRGLEERCLCLSACEASEEEL
GLVHSPEYIALVQKTQTLDKEELHALSKQYNAVYFHPDTFHCARLAAGAALQLVDAVLTG
AVHNGLALVRPPGHHSQRAAANGFCVFNNVALAAKHAKQKYGLQRILIVDWDVHHGQGIQ
YIFNDDPSVLYFSWHRYEHGSFWPFLPESDADAVGQGQGQGFTVNLPWNQVGMGNADYLA
AFLHVLLPLAFEFDPELVLVSAGFDSAIGDPEGQMQATPECFAHLTQLLQVLAGGRICAV
LEGGYHLESLAQSVCMMVQTLLGDPTPPLLGLMVPCQSALESIQSVQTAQTPYWTSLQQN
VAPVLSSSTHSPEERSLRLLGESPTCAVAEDSLSPLLDQLCLRPAPPICTAVASTVPGAA
LCLPPGVLHQEGSVLREETEAWARLHKSRFQDEDLATLGKILCLLDGIMDGQIRNAIATT
TALATAATLDVLIQRCLARRAQRVLCVALGQLDRPLDLADDGRILWLNIRGKDAAIQSMF
HFSTPLPQTTGGFLSLILGLVLPLAYGFQPDMVLMALGPAHGLQNAQAALLAAMLRSPVG
GRILAVVEEESIRLLARSLAQALHGETPPSLGPFSKATPEEIQALMFLKARLEARWKLLQ
VAAPPP
|
|
|
|---|
| BDBM50366962 |
|---|
| n/a |
|---|
| Name | BDBM50366962 |
|---|
| Synonyms: | CHEMBL1790586 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C30H45N5O7 |
|---|
| Mol. Mass. | 587.7076 |
|---|
| SMILES | CC[C@@H](C)[C@@H]1NC(=O)[C@@H](Cc2ccc(OC)cc2)NC(=O)[C@H](CCCCCN(O)C=O)NC(=O)[C@H]2CCCCN2C1=O |
|---|
| Structure | 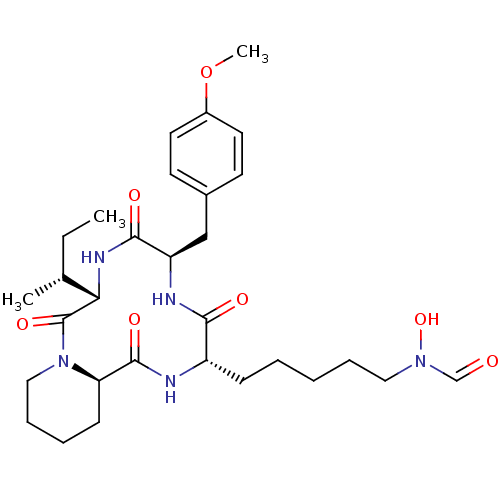
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nishino, N; Yoshikawa, D; Watanabe, LA; Kato, T; Jose, B; Komatsu, Y; Sumida, Y; Yoshida, M Synthesis and histone deacetylase inhibitory activity of cyclic tetrapeptides containing a retrohydroxamate as zinc ligand. Bioorg Med Chem Lett14:2427-31 (2004) [PubMed] Article
Nishino, N; Yoshikawa, D; Watanabe, LA; Kato, T; Jose, B; Komatsu, Y; Sumida, Y; Yoshida, M Synthesis and histone deacetylase inhibitory activity of cyclic tetrapeptides containing a retrohydroxamate as zinc ligand. Bioorg Med Chem Lett14:2427-31 (2004) [PubMed] Article