| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Ligand | BDBM50452435 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_53174 |
|---|
| Ki | 30±n/a nM |
|---|
| Citation |  Baldwin, JJ; Claremon, DA; Lumma, PK; McClure, DE; Rosenthal, SA; Winquist, RJ; Faison, EP; Kaczorowski, GJ; Trumble, MJ; Smith, GM Diethyl 3,6-dihydro-2,4-dimethyl-2,6-methano-1,3-benzothiazocine-5,11- dicarboxylates as calcium entry antagonists: new conformationally restrained analogues of Hantzsch 1,4-dihydropyridines related to nitrendipine as probes for receptor-site conformation. J Med Chem30:690-5 (1987) [PubMed] Baldwin, JJ; Claremon, DA; Lumma, PK; McClure, DE; Rosenthal, SA; Winquist, RJ; Faison, EP; Kaczorowski, GJ; Trumble, MJ; Smith, GM Diethyl 3,6-dihydro-2,4-dimethyl-2,6-methano-1,3-benzothiazocine-5,11- dicarboxylates as calcium entry antagonists: new conformationally restrained analogues of Hantzsch 1,4-dihydropyridines related to nitrendipine as probes for receptor-site conformation. J Med Chem30:690-5 (1987) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Name: | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Synonyms: | CAC1C_HUMAN | CACH2 | CACN2 | CACNA1C | CACNL1A1 | CCHL1A1 | Calcium channel (Type L) | Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle | L-type calcium channel alpha-1c/beta-2/alpha2delta-1 | Voltage-dependent L-type calcium channel subunit alpha-1C | Voltage-gated L-type calcium channel | Voltage-gated L-type calcium channel alpha-1C subunit | Voltage-gated calcium channel | Voltage-gated calcium channel subunit alpha Cav1.2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 248979.79 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Calcium channel (Type L) 0 HUMAN::Q13936 |
|---|
| Residue: | 2221 |
|---|
| Sequence: | MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAAR
QAKLMGSAGNATISTVSSTQRKRQQYGKPKKQGSTTATRPPRALLCLTLKNPIRRACISI
VEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA
YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFR
VLRPLRLVSGVPSLQVVLNSIIKAMVPLLHIALLVLFVIIIYAIIGLELFMGKMHKTCYN
QEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI
TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGD
FQKLREKQQLEEDLKGYLDWITQAEDIDPENEDEGMDEEKPRNMSMPTSETESVNTENVA
GGDIEGENCGARLAHRISKSKFSRYWRRWNRFCRRKCRAAVKSNVFYWLVIFLVFLNTLT
IASEHYNQPNWLTEVQDTANKALLALFTAEMLLKMYSLGLQAYFVSLFNRFDCFVVCGGI
LETILVETKIMSPLGISVLRCVRLLRIFKITRYWNSLSNLVASLLNSVRSIASLLLLLFL
FIIIFSLLGMQLFGGKFNFDEMQTRRSTFDNFPQSLLTVFQILTGEDWNSVMYDGIMAYG
GPSFPGMLVCIYFIILFICGNYILLNVFLAIAVDNLADAESLTSAQKEEEEEKERKKLAR
TASPEKKQELVEKPAVGESKEEKIELKSITADGESPPATKINMDDLQPNENEDKSPYPNP
ETTGEEDEEEPEMPVGPRPRPLSELHLKEKAVPMPEASAFFIFSSNNRFRLQCHRIVNDT
IFTNLILFFILLSSISLAAEDPVQHTSFRNHILFYFDIVFTTIFTIEIALKILGNADYVF
TSIFTLEIILKMTAYGAFLHKGSFCRNYFNILDLLVVSVSLISFGIQSSAINVVKILRVL
RVLRPLRAINRAKGLKHVVQCVFVAIRTIGNIVIVTTLLQFMFACIGVQLFKGKLYTCSD
SSKQTEAECKGNYITYKDGEVDHPIIQPRSWENSKFDFDNVLAAMMALFTVSTFEGWPEL
LYRSIDSHTEDKGPIYNYRVEISIFFIIYIIIIAFFMMNIFVGFVIVTFQEQGEQEYKNC
ELDKNQRQCVEYALKARPLRRYIPKNQHQYKVWYVVNSTYFEYLMFVLILLNTICLAMQH
YGQSCLFKIAMNILNMLFTGLFTVEMILKLIAFKPKGYFSDPWNVFDFLIVIGSIIDVIL
SETNHYFCDAWNTFDALIVVGSIVDIAITEVNPAEHTQCSPSMNAEENSRISITFFRLFR
VMRLVKLLSRGEGIRTLLWTFIKSFQALPYVALLIVMLFFIYAVIGMQVFGKIALNDTTE
INRNNNFQTFPQAVLLLFRCATGEAWQDIMLACMPGKKCAPESEPSNSTEGETPCGSSFA
VFYFISFYMLCAFLIINLFVAVIMDNFDYLTRDWSILGPHHLDEFKRIWAEYDPEAKGRI
KHLDVVTLLRRIQPPLGFGKLCPHRVACKRLVSMNMPLNSDGTVMFNATLFALVRTALRI
KTEGNLEQANEELRAIIKKIWKRTSMKLLDQVVPPAGDDEVTVGKFYATFLIQEYFRKFK
KRKEQGLVGKPSQRNALSLQAGLRTLHDIGPEIRRAISGDLTAEEELDKAMKEAVSAASE
DDIFRRAGGLFGNHVSYYQSDGRSAFPQTFTTQRPLHINKAGSSQGDTESPSHEKLVDST
FTPSSYSSTGSNANINNANNTALGRLPRPAGYPSTVSTVEGHGPPLSPAIRVQEVAWKLS
SNRERHVPMCEDLELRRDSGSAGTQAHCLLLRKANPSRCHSRESQAAMAGQEETSQDETY
EVKMNHDTEACSEPSLLSTEMLSYQDDENRQLTLPEEDKRDIRQSPKRGFLRSASLGRRA
SFHLECLKRQKDRGGDISQKTVLPLHLVHHQALAVAGLSPLLQRSHSPASFPRPFATPPA
TPGSRGWPPQPVPTLRLEGVESSEKLNSSFPSIHCGSWAETTPGGGGSSAARRVRPVSLM
VPSQAGAPGRQFHGSASSLVEAVLISEGLGQFAQDPKFIEVTTQELADACDMTIEEMESA
ADNILSGGAPQSPNGALLPFVNCRDAGQDRAGGEEDAGCVRARGRPSEEELQDSRVYVSS
L
|
|
|
|---|
| BDBM50452435 |
|---|
| n/a |
|---|
| Name | BDBM50452435 |
|---|
| Synonyms: | CHEMBL2110302 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H22N2O6S |
|---|
| Mol. Mass. | 406.453 |
|---|
| SMILES | [H][C@@]12[C@@H](C(=O)OCC)[C@@](C)(Sc3ccc(cc13)[N+]([O-])=O)N=C(C)\C2=C(\O)OCC |t:22,THB:12:11:2:23.21.20,15:16:2:23.21.20,24:23:2:16.10.11| |
|---|
| Structure | 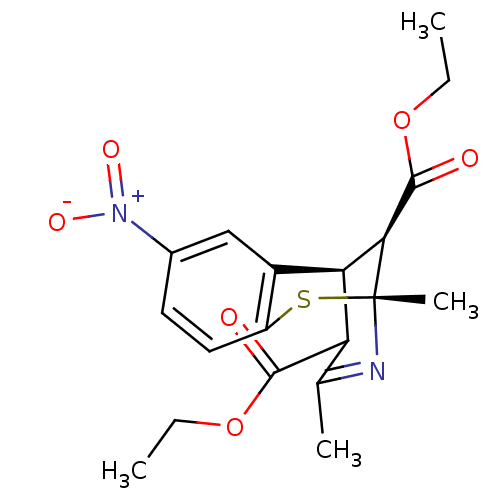
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Baldwin, JJ; Claremon, DA; Lumma, PK; McClure, DE; Rosenthal, SA; Winquist, RJ; Faison, EP; Kaczorowski, GJ; Trumble, MJ; Smith, GM Diethyl 3,6-dihydro-2,4-dimethyl-2,6-methano-1,3-benzothiazocine-5,11- dicarboxylates as calcium entry antagonists: new conformationally restrained analogues of Hantzsch 1,4-dihydropyridines related to nitrendipine as probes for receptor-site conformation. J Med Chem30:690-5 (1987) [PubMed]
Baldwin, JJ; Claremon, DA; Lumma, PK; McClure, DE; Rosenthal, SA; Winquist, RJ; Faison, EP; Kaczorowski, GJ; Trumble, MJ; Smith, GM Diethyl 3,6-dihydro-2,4-dimethyl-2,6-methano-1,3-benzothiazocine-5,11- dicarboxylates as calcium entry antagonists: new conformationally restrained analogues of Hantzsch 1,4-dihydropyridines related to nitrendipine as probes for receptor-site conformation. J Med Chem30:690-5 (1987) [PubMed]