| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Aminopeptidase N |
|---|
| Ligand | BDBM50036828 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_35505 (CHEMBL646415) |
|---|
| IC50 | 30±n/a nM |
|---|
| Citation |  Chauvel, EN; Coric, P; Llorens-Cortès, C; Wilk, S; Roques, BP; Fournié-Zaluski, MC Investigation of the active site of aminopeptidase A using a series of new thiol-containing inhibitors. J Med Chem37:1339-46 (1994) [PubMed] Chauvel, EN; Coric, P; Llorens-Cortès, C; Wilk, S; Roques, BP; Fournié-Zaluski, MC Investigation of the active site of aminopeptidase A using a series of new thiol-containing inhibitors. J Med Chem37:1339-46 (1994) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Aminopeptidase N |
|---|
| Name: | Aminopeptidase N |
|---|
| Synonyms: | AMPN_HUMAN | ANPEP | APN | Alanyl aminopeptidase | Aminopeptidase | CD13 | CD_antigen=CD13 | Microsomal aminopeptidase | Myeloid plasma membrane glycoprotein CD13 | PEPN | gp150 | hAPN |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 109522.63 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1507526 |
|---|
| Residue: | 967 |
|---|
| Sequence: | MAKGFYISKSLGILGILLGVAAVCTIIALSVVYSQEKNKNANSSPVASTTPSASATTNPA
SATTLDQSKAWNRYRLPNTLKPDSYRVTLRPYLTPNDRGLYVFKGSSTVRFTCKEATDVI
IIHSKKLNYTLSQGHRVVLRGVGGSQPPDIDKTELVEPTEYLVVHLKGSLVKDSQYEMDS
EFEGELADDLAGFYRSEYMEGNVRKVVATTQMQAADARKSFPCFDEPAMKAEFNITLIHP
KDLTALSNMLPKGPSTPLPEDPNWNVTEFHTTPKMSTYLLAFIVSEFDYVEKQASNGVLI
RIWARPSAIAAGHGDYALNVTGPILNFFAGHYDTPYPLPKSDQIGLPDFNAGAMENWGLV
TYRENSLLFDPLSSSSSNKERVVTVIAHELAHQWFGNLVTIEWWNDLWLNEGFASYVEYL
GADYAEPTWNLKDLMVLNDVYRVMAVDALASSHPLSTPASEINTPAQISELFDAISYSKG
ASVLRMLSSFLSEDVFKQGLASYLHTFAYQNTIYLNLWDHLQEAVNNRSIQLPTTVRDIM
NRWTLQMGFPVITVDTSTGTLSQEHFLLDPDSNVTRPSEFNYVWIVPITSIRDGRQQQDY
WLIDVRAQNDLFSTSGNEWVLLNLNVTGYYRVNYDEENWRKIQTQLQRDHSAIPVINRAQ
IINDAFNLASAHKVPVTLALNNTLFLIEERQYMPWEAALSSLSYFKLMFDRSEVYGPMKN
YLKKQVTPLFIHFRNNTNNWREIPENLMDQYSEVNAISTACSNGVPECEEMVSGLFKQWM
ENPNNNPIHPNLRSTVYCNAIAQGGEEEWDFAWEQFRNATLVNEADKLRAALACSKELWI
LNRYLSYTLNPDLIRKQDATSTIISITNNVIGQGLVWDFVQSNWKKLFNDYGGGSFSFSN
LIQAVTRRFSTEYELQQLEQFKKDNEETGFGSGTRALEQALEKTKANIKWVKENKEVVLQ
WFTENSK
|
|
|
|---|
| BDBM50036828 |
|---|
| n/a |
|---|
| Name | BDBM50036828 |
|---|
| Synonyms: | 1-Mercaptomethyl-2-phenyl-ethyl-ammonium | 2-Amino-3-phenyl-propane-1-thiol | CHEMBL414393 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C9H13NS |
|---|
| Mol. Mass. | 167.271 |
|---|
| SMILES | NC(CS)Cc1ccccc1 |
|---|
| Structure | 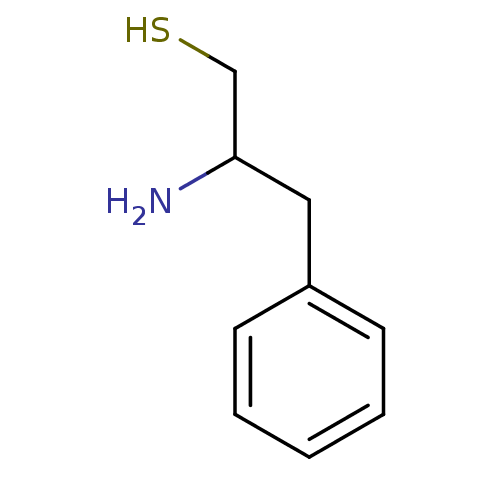
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chauvel, EN; Coric, P; Llorens-Cortès, C; Wilk, S; Roques, BP; Fournié-Zaluski, MC Investigation of the active site of aminopeptidase A using a series of new thiol-containing inhibitors. J Med Chem37:1339-46 (1994) [PubMed]
Chauvel, EN; Coric, P; Llorens-Cortès, C; Wilk, S; Roques, BP; Fournié-Zaluski, MC Investigation of the active site of aminopeptidase A using a series of new thiol-containing inhibitors. J Med Chem37:1339-46 (1994) [PubMed]