| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Maltase-glucoamylase |
|---|
| Ligand | BDBM50259956 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_34390 (CHEMBL649260) |
|---|
| IC50 | 40±n/a nM |
|---|
| Citation |  Asano, N; Nishida, M; Kato, A; Kizu, H; Matsui, K; Shimada, Y; Itoh, T; Baba, M; Watson, AA; Nash, RJ; Lilley, PM; Watkin, DJ; Fleet, GW Homonojirimycin isomers and N-alkylated homonojirimycins: structural and conformational basis of inhibition of glycosidases. J Med Chem41:2565-71 (1998) [PubMed] Article Asano, N; Nishida, M; Kato, A; Kizu, H; Matsui, K; Shimada, Y; Itoh, T; Baba, M; Watson, AA; Nash, RJ; Lilley, PM; Watkin, DJ; Fleet, GW Homonojirimycin isomers and N-alkylated homonojirimycins: structural and conformational basis of inhibition of glycosidases. J Med Chem41:2565-71 (1998) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Maltase-glucoamylase |
|---|
| Name: | Maltase-glucoamylase |
|---|
| Synonyms: | Alpha glucosidase | Alpha-1,4-glucosidase | Glucan 1,4-alpha-glucosidase | MGA | MGAM | MGAML | MGA_HUMAN | Maltase | Maltase-glucoamylase, intestinal | Synonyms=MGA |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 209817.06 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O43451 |
|---|
| Residue: | 2753 |
|---|
| Sequence: | MARKKLKKFTTLEIVLSVLLLVLFIISIVLIVLLAKESLKSTAPDPGTTGTPDPGTTGTP
DPGTTGTTHARTTGPPDPGTTGTTPVSAECPVVNELERINCIPDQPPTKATCDQRGCCWN
PQGAVSVPWCYYSKNHSYHVEGNLVNTNAGFTARLKNLPSSPVFGSNVDNVLLTAEYQTS
NRFHFKLTDQTNNRFEVPHEHVQSFSGNAAASLTYQVEISRQPFSIKVTRRSNNRVLFDS
SIGPLLFADQFLQLSTRLPSTNVYGLGEHVHQQYRHDMNWKTWPIFNRDTTPNGNGTNLY
GAQTFFLCLEDASGLSFGVFLMNSNAMEVVLQPAPAITYRTIGGILDFYVFLGNTPEQVV
QEYLELIGRPALPSYWALGFHLSRYEYGTLDNMREVVERNRAAQLPYDVQHADIDYMDER
RDFTYDSVDFKGFPEFVNELHNNGQKLVIIVDPAISNNSSSSKPYGPYDRGSDMKIWVNS
SDGVTPLIGEVWPGQTVFPDYTNPNCAVWWTKEFELFHNQVEFDGIWIDMNEVSNFVDGS
VSGCSTNNLNNPPFTPRILDGYLFCKTLCMDAVQHWGKQYDIHNLYGYSMAVATAEAAKT
VFPNKRSFILTRSTFAGSGKFAAHWLGDNTATWDDLRWSIPGVLEFNLFGIPMVGPDICG
FALDTPEELCRRWMQLGAFYPFSRNHNGQGYKDQDPASFGADSLLLNSSRHYLNIRYTLL
PYLYTLFFRAHSRGDTVARPLLHEFYEDNSTWDVHQQFLWGPGLLITPVLDEGAEKVMAY
VPDAVWYDYETGSQVRWRKQKVEMELPGDKIGLHLRGGYIFPTQQPNTTTLASRKNPLGL
IIALDENKEAKGELFWDNGETKDTVANKVYLLCEFSVTQNRLEVNISQSTYKDPNNLAFN
EIKILGTEEPSNVTVKHNGVPSQTSPTVTYDSNLKVAIITDIDLLLGEAYTVEWSIKIRD
EEKIDCYPDENGASAENCTARGCIWEASNSSGVPFCYFVNDLYSVSDVQYNSHGATADIS
LKSSVYANAFPSTPVNPLRLDVTYHKNEMLQFKIYDPNKNRYEVPVPLNIPSMPSSTPEG
QLYDVLIKKNPFGIEIRRKSTGTIIWDSQLLGFTFSDMFIRISTRLPSKYLYGFGETEHR
SYRRDLEWHTWGMFSRDQPPGYKKNSYGVHPYYMGLEEDGSAHGVLLLNSNAMDVTFQPL
PALTYRTTGGVLDFYVFLGPTPELVTQQYTELIGRPVMVPYWSLGFQLCRYGYQNDSEIA
SLYDEMVAAQIPYDVQYSDIDYMERQLDFTLSPKFAGFPALINRMKADGMRVILILDPAI
SGNETQPYPAFTRGVEDDVFIKYPNDGDIVWGKVWPDFPDVVVNGSLDWDSQVELYRAYV
AFPDFFRNSTAKWWKREIEELYNNPQNPERSLKFDGMWIDMNEPSSFVNGAVSPGCRDAS
LNHPPYMPHLESRDRGLSSKTLCMESQQILPDGSLVQHYNVHNLYGWSQTRPTYEAVQEV
TGQRGVVITRSTFPSSGRWAGHWLGDNTAAWDQLKKSIIGMMEFSLFGISYTGADICGFF
QDAEYEMCVRWMQLGAFYPFSRNHNTIGTRRQDPVSWDAAFVNISRNVLQTRYTLLPYLY
TLMQKAHTEGVTVVRPLLHEFVSDQVTWDIDSQFLLGPAFLVSPVLERNARNVTAYFPRA
RWYDYYTGVDINARGEWKTLPAPLDHINLHVRGGYILPWQEPALNTHLSRKNPLGLIIAL
DENKEAKGELFWDDGQTKDTVAKKVYLLCEFSVTQNHLEVTISQSTYKDPNNLAFNEIKI
LGMEEPSNVTVKHNGVPSQTSPTVTYDSNLKVAIITDINLFLGEAYTVEWSIKIRDEEKI
DCYPDENGDSAENCTARGCIWEASNSSGVPFCYFVNDLYSVSDVQYNSHGATADISLKSS
VHANAFPSTPVNPLRLDVTYHKNEMLQFKIYDPNNNRYEVPVPLNIPSVPSSTPEGQLYD
VLIKKNPFGIEIRRKSTGTIIWDSQLLGFTFNDMFIRISTRLPSKYLYGFGETEHTSYRR
DLEWHTWGMFSRDQPPGYKKNSYGVHPYYMGLEEDGSAHGVLLLNSNAMDVTFQPLPALT
YRTTGGVLDFYVFLGPTPELVTQQYTELIGRPVMVPYWSLGFQLCRYGYQNDSEISSLYD
EMVAAQIPYDVQYSDIDYMERQLDFTLSPKFAGFPALINRMKADGMRVILILDPAISGNE
TQPYPAFTRGVEDDVFIKYPNDGDIVWGKVWPDFPDVVVNGSLDWDSQVELYRAYVAFPD
FFRNSTAKWWKREIEELYNNPQNPERSLKFDGMWIDMNEPSSFVNGAVSPGCRDASLNHP
PYMPYLESRDRGLSSKTLCMESQQILPDGSPVQHYNVHNLYGWSQTRPTYEAVQEVTGQR
GVVITRSTFPSSGRWAGHWLGDNTAAWDQLKKSIIGMMEFSLFGISYTGADICGFFQDAE
YEMCVRWMQLGAFYPFSRNHNTIGTRRQDPVSWDVAFVNISRTVLQTRYTLLPYLYTLMH
KAHTEGVTVVRPLLHEFVSDQVTWDIDSQFLLGPAFLVSPVLERNARNVTAYFPRARWYD
YYTGVDINARGEWKTLPAPLDHINLHVRGGYILPWQEPALNTHLSRQKFMGFKIALDDEG
TAGGWLFWDDGQSIDTYGKGLYYLASFSASQNTMQSHIIFNNYITGTNPLKLGYIEIWGV
GSVPVTSVSISVSGMVITPSFNNDPTTQVLSIDVTDRNISLHNFTSLTWISTL
|
|
|
|---|
| BDBM50259956 |
|---|
| n/a |
|---|
| Name | BDBM50259956 |
|---|
| Synonyms: | 2,6-Bis-hydroxymethyl-piperidine-3,4,5-triol | CHEMBL501355 | US9181184, 34 | alpha-homonojirimycin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C7H15NO5 |
|---|
| Mol. Mass. | 193.1977 |
|---|
| SMILES | OC[C@H]1N[C@H](CO)[C@H](O)[C@@H](O)[C@@H]1O |r| |
|---|
| Structure | 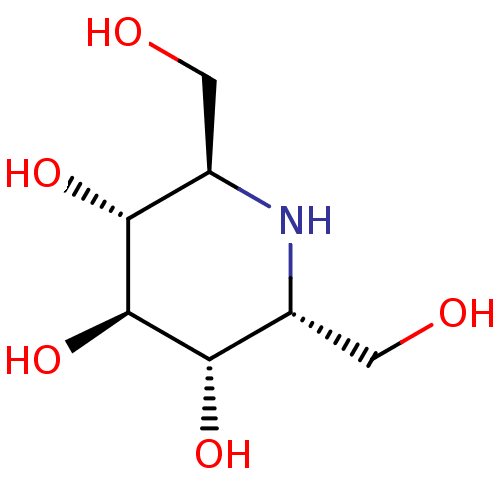
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Asano, N; Nishida, M; Kato, A; Kizu, H; Matsui, K; Shimada, Y; Itoh, T; Baba, M; Watson, AA; Nash, RJ; Lilley, PM; Watkin, DJ; Fleet, GW Homonojirimycin isomers and N-alkylated homonojirimycins: structural and conformational basis of inhibition of glycosidases. J Med Chem41:2565-71 (1998) [PubMed] Article
Asano, N; Nishida, M; Kato, A; Kizu, H; Matsui, K; Shimada, Y; Itoh, T; Baba, M; Watson, AA; Nash, RJ; Lilley, PM; Watkin, DJ; Fleet, GW Homonojirimycin isomers and N-alkylated homonojirimycins: structural and conformational basis of inhibition of glycosidases. J Med Chem41:2565-71 (1998) [PubMed] Article