| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-lactamase |
|---|
| Ligand | BDBM50067069 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_41032 (CHEMBL651936) |
|---|
| IC50 | 26±n/a nM |
|---|
| Citation |  Hubschwerlen, C; Angehrn, P; Gubernator, K; Page, MG; Specklin, JL Structure-based design of beta-lactamase inhibitors. 2. Synthesis and evaluation of bridged sulfactams and oxamazins. J Med Chem41:3972-5 (1998) [PubMed] Article Hubschwerlen, C; Angehrn, P; Gubernator, K; Page, MG; Specklin, JL Structure-based design of beta-lactamase inhibitors. 2. Synthesis and evaluation of bridged sulfactams and oxamazins. J Med Chem41:3972-5 (1998) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-lactamase |
|---|
| Name: | Beta-lactamase |
|---|
| Synonyms: | AMPC_ENTCL | ampC |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 41306.67 |
|---|
| Organism: | Enterobacter cloacae |
|---|
| Description: | ChEMBL_40258 |
|---|
| Residue: | 381 |
|---|
| Sequence: | MMRKSLCCALLLGISCSALATPVSEKQLAEVVANTITPLMKAQSVPGMAVAVIYQGKPHY
YTFGKADIAANKPVTPQTLFELGSISKTFTGVLGGDAIARGEISLDDAVTRYWPQLTGKQ
WQGIRMLDLATYTAGGLPLQVPDEVTDNASLLRFYQNWQPQWKPGTTRLYANASIGLFGA
LAVKPSGMPYEQAMTTRVLKPLKLDHTWINVPKAEEAHYAWGYRDGKAVRVSPGMLDAQA
YGVKTNVQDMANWVMANMAPENVADASLKQGIALAQSRYWRIGSMYQGLGWEMLNWPVEA
NTVVEGSDSKVALAPLPVAEVNPPAPPVKASWVHKTGSTGGFGSYVAFIPEKQIGIVMLA
NTSYPNPARVEAAYHILEALQ
|
|
|
|---|
| BDBM50067069 |
|---|
| n/a |
|---|
| Name | BDBM50067069 |
|---|
| Synonyms: | CHEMBL128220 | sodium salt-Sulfuric acid mono-[2-(4-carbamoyl-phenylcarbamoyl)-7-oxo-2,6-diaza-bicyclo[3.2.0]hept-6-yl] ester |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H13N4O7S |
|---|
| Mol. Mass. | 369.33 |
|---|
| SMILES | NC(=O)c1ccc(NC(=O)N2CCC3C2C(=O)N3OS([O-])(=O)=O)cc1 |
|---|
| Structure | 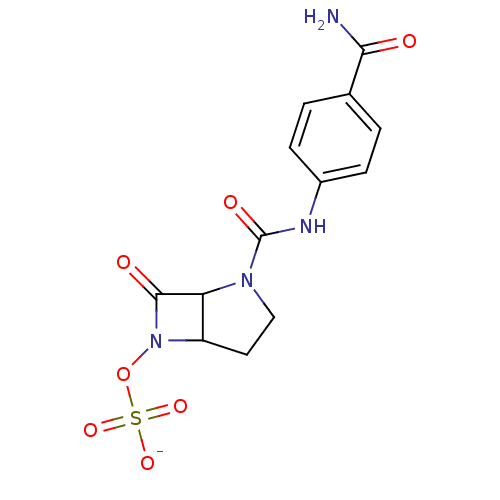
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hubschwerlen, C; Angehrn, P; Gubernator, K; Page, MG; Specklin, JL Structure-based design of beta-lactamase inhibitors. 2. Synthesis and evaluation of bridged sulfactams and oxamazins. J Med Chem41:3972-5 (1998) [PubMed] Article
Hubschwerlen, C; Angehrn, P; Gubernator, K; Page, MG; Specklin, JL Structure-based design of beta-lactamase inhibitors. 2. Synthesis and evaluation of bridged sulfactams and oxamazins. J Med Chem41:3972-5 (1998) [PubMed] Article