| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 5-hydroxytryptamine receptor 3A/3B |
|---|
| Ligand | BDBM50053631 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_3233 (CHEMBL618919) |
|---|
| Ki | 3±n/a nM |
|---|
| Citation |  Cappelli, A; Anzini, M; Vomero, S; Canullo, L; Mennuni, L; Makovec, F; Doucet, E; Hamon, M; Menziani, MC; De Benedetti, PG; Bruni, G; Romeo, MR; Giorgi, G; Donati, A Novel potent and selective central 5-HT3 receptor ligands provided with different intrinsic efficacy. 2. Molecular basis of the intrinsic efficacy of arylpiperazine derivatives at the central 5-HT3 receptors. J Med Chem42:1556-75 (1999) [PubMed] Article Cappelli, A; Anzini, M; Vomero, S; Canullo, L; Mennuni, L; Makovec, F; Doucet, E; Hamon, M; Menziani, MC; De Benedetti, PG; Bruni, G; Romeo, MR; Giorgi, G; Donati, A Novel potent and selective central 5-HT3 receptor ligands provided with different intrinsic efficacy. 2. Molecular basis of the intrinsic efficacy of arylpiperazine derivatives at the central 5-HT3 receptors. J Med Chem42:1556-75 (1999) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 5-hydroxytryptamine receptor 3A/3B |
|---|
| Name: | 5-hydroxytryptamine receptor 3A/3B |
|---|
| Synonyms: | Serotonin 3 (5-HT3) receptor | Serotonin 3a (5-HT3a)/3b (5-HT3b) receptor |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of EBI is 2974 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | 5-hydroxytryptamine receptor 3B |
|---|
| Synonyms: | 5HT3B_RAT | Htr3b | Serotonin (5-HT) receptor |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 50328.78 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | EBI_11885 |
|---|
| Residue: | 437 |
|---|
| Sequence: | MILLWSCLLVAVVGILGTATPQPGNSSLHRLTRQLLQQYHKEVRPVYNWAEATTVYLDLC
VHAVLDVDVQNQKLKTSMWYREVWNDEFLSWNSSLFDDIQEISLPLSAIWAPDIIINEFV
DVERSPDLPYVYVNSSGTIRNHKPIQVVSACSLQTYAFPFDIQNCSLTFNSILHTVEDID
LGFLRNQEDIENDKRSFLNDSEWQLLSVTSTYHIRQSSAGDFAQIRFNVVIRRCPLAYVV
SLLIPSIFLMLVDLGSFYLPPNCRARIVFKTNVLVGYTVFRVNMSDEVPRSAGCTSLIGV
FFTVCMALLVLSLSKSILLIKFLYEERHSEQERPLMCLRGDSDANESRLYLRAPCAEDTE
SPVRQEHQVPSDTLKDFWFQLQSINNSLRTRDQVYQKEVEWLAILCHFDQLLFRIYLAVL
GLYTVTLCSLWALWSRM
|
|
|
|---|
| Component 2 |
| Name: | 5-hydroxytryptamine receptor 3A |
|---|
| Synonyms: | 5-HT3 | 5-hydroxytryptamine receptor 3A | 5HT3A_RAT | 5ht3 | Htr3 | Htr3a | Serotonin (5-HT) receptor | Zacopride site-R |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 55428.70 |
|---|
| Organism: | RAT |
|---|
| Description: | 5-HT3 HTR3A RAT::P35563 |
|---|
| Residue: | 483 |
|---|
| Sequence: | MPLCIPQVLLALFLSVLIAQGEGSRRRATQAHSTTQPALLRLSDHLLANYKKGVRPVRDW
RKPTLVSIDVIMYAILNVDEKNQVLTTYIWYRQFWTDEFLQWTPEDFDNVTKLSIPTDSI
WVPDILINEFVDVGKSPSIPYVYVHHQGEVQNYKPLQLVTACSLDIYNFPFDVQNCSLTF
TSWLHTIQDINISLWRTPEEVRSDKSIFINQGEWELLGVFTKFQEFSIETSNSYAEMKFY
VVIRRRPLFYAVSLLLPSIFLMVVDIVGFCLPPDSGERVSFKITLLLGYSVFLIIVSDTL
PATAIGTPLIGVYFVVCMALLVISLAETIFIVQLVHKQDLQRPVPDWLRHLVLDRIAWLL
CLGEQPMAHRPPATFQANKTDDCSAMGNHCSHVGSPQDLEKTSRSRDSPLPPPREASLAV
RGLLQELSSIRHSLEKRDEMREVARDWLRVGYVLDRLLFRIYLLAVLAYSITLVTLWSIW
HYS
|
|
|
|---|
| BDBM50053631 |
|---|
| n/a |
|---|
| Name | BDBM50053631 |
|---|
| Synonyms: | 2-(4-Methyl-piperazin-1-yl)-quinoline | 2-(4-methylpiperazin-1-yl)quinoline | CHEMBL288591 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H17N3 |
|---|
| Mol. Mass. | 227.3049 |
|---|
| SMILES | CN1CCN(CC1)c1ccc2ccccc2n1 |
|---|
| Structure | 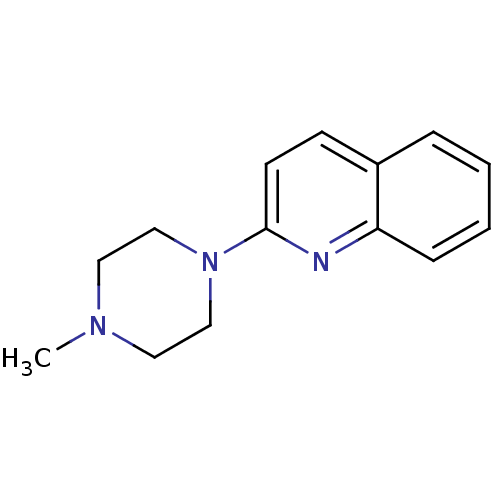
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cappelli, A; Anzini, M; Vomero, S; Canullo, L; Mennuni, L; Makovec, F; Doucet, E; Hamon, M; Menziani, MC; De Benedetti, PG; Bruni, G; Romeo, MR; Giorgi, G; Donati, A Novel potent and selective central 5-HT3 receptor ligands provided with different intrinsic efficacy. 2. Molecular basis of the intrinsic efficacy of arylpiperazine derivatives at the central 5-HT3 receptors. J Med Chem42:1556-75 (1999) [PubMed] Article
Cappelli, A; Anzini, M; Vomero, S; Canullo, L; Mennuni, L; Makovec, F; Doucet, E; Hamon, M; Menziani, MC; De Benedetti, PG; Bruni, G; Romeo, MR; Giorgi, G; Donati, A Novel potent and selective central 5-HT3 receptor ligands provided with different intrinsic efficacy. 2. Molecular basis of the intrinsic efficacy of arylpiperazine derivatives at the central 5-HT3 receptors. J Med Chem42:1556-75 (1999) [PubMed] Article