| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM50113849 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_212197 (CHEMBL817760) |
|---|
| Ki | 1200±n/a nM |
|---|
| Citation |  Cozzini, P; Fornabaio, M; Marabotti, A; Abraham, DJ; Kellogg, GE; Mozzarelli, A Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J Med Chem45:2469-83 (2002) [PubMed] Cozzini, P; Fornabaio, M; Marabotti, A; Abraham, DJ; Kellogg, GE; Mozzarelli, A Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J Med Chem45:2469-83 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Alpha-trypsin chain 1 | Alpha-trypsin chain 2 | Beta-trypsin | Cationic trypsinogen | PRSS1 | Serine protease 1 | TRP1 | TRY1 | TRY1_HUMAN | TRYP1 | Thrombin & trypsin | Trypsin | Trypsin I | Trypsin-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 26557.80 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P07477 |
|---|
| Residue: | 247 |
|---|
| Sequence: | MNPLLILTFVAAALAAPFDDDDKIVGGYNCEENSVPYQVSLNSGYHFCGGSLINEQWVVS
AGHCYKSRIQVRLGEHNIEVLEGNEQFINAAKIIRHPQYDRKTLNNDIMLIKLSSRAVIN
ARVSTISLPTAPPATGTKCLISGWGNTASSGADYPDELQCLDAPVLSQAKCEASYPGKIT
SNMFCVGFLEGGKDSCQGDSGGPVVCNGQLQGVVSWGDGCAQKNKPGVYTKVYNYVKWIK
NTIAANS
|
|
|
|---|
| BDBM50113849 |
|---|
| n/a |
|---|
| Name | BDBM50113849 |
|---|
| Synonyms: | 3-[3-Oxo-3-piperidin-1-yl-2-(toluene-4-sulfonylamino)-propyl]-benzamidine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H29N4O3S |
|---|
| Mol. Mass. | 429.555 |
|---|
| SMILES | Cc1ccc(cc1)S(=O)(=O)NC(Cc1cccc(c1)C(N)=[NH2+])C(=O)N1CCCCC1 |
|---|
| Structure | 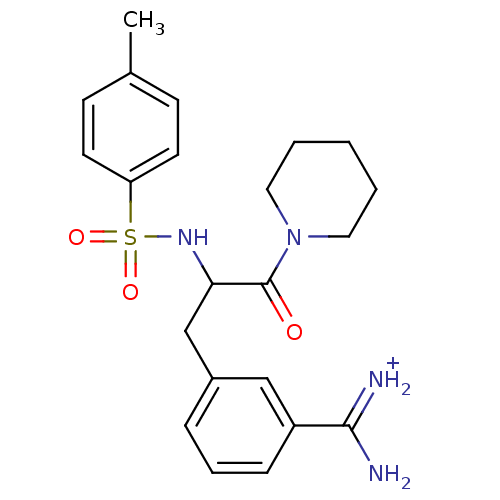
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cozzini, P; Fornabaio, M; Marabotti, A; Abraham, DJ; Kellogg, GE; Mozzarelli, A Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J Med Chem45:2469-83 (2002) [PubMed]
Cozzini, P; Fornabaio, M; Marabotti, A; Abraham, DJ; Kellogg, GE; Mozzarelli, A Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J Med Chem45:2469-83 (2002) [PubMed]