| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor 1 |
|---|
| Ligand | BDBM50207594 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_90140 (CHEMBL701239) |
|---|
| Ki | 1600±n/a nM |
|---|
| Citation |  Filla, SA; Winter, MA; Johnson, KW; Bleakman, D; Bell, MG; Bleisch, TJ; Castaño, AM; Clemens-Smith, A; del Prado, M; Dieckman, DK; Dominguez, E; Escribano, A; Ho, KH; Hudziak, KJ; Katofiasc, MA; Martinez-Perez, JA; Mateo, A; Mathes, BM; Mattiuz, EL; Ogden, AM; Phebus, LA; Stack, DR; Stratford, RE; Ornstein, PL Ethyl (3S,4aR,6S,8aR)-6-(4-ethoxycar- bonylimidazol-1-ylmethyl)decahydroiso-quinoline-3-carboxylic ester: a prodrug of a GluR5 kainate receptor antagonist active in two animal models of acute migraine. J Med Chem45:4383-6 (2002) [PubMed] Filla, SA; Winter, MA; Johnson, KW; Bleakman, D; Bell, MG; Bleisch, TJ; Castaño, AM; Clemens-Smith, A; del Prado, M; Dieckman, DK; Dominguez, E; Escribano, A; Ho, KH; Hudziak, KJ; Katofiasc, MA; Martinez-Perez, JA; Mateo, A; Mathes, BM; Mattiuz, EL; Ogden, AM; Phebus, LA; Stack, DR; Stratford, RE; Ornstein, PL Ethyl (3S,4aR,6S,8aR)-6-(4-ethoxycar- bonylimidazol-1-ylmethyl)decahydroiso-quinoline-3-carboxylic ester: a prodrug of a GluR5 kainate receptor antagonist active in two animal models of acute migraine. J Med Chem45:4383-6 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate receptor 1 |
|---|
| Name: | Glutamate receptor 1 |
|---|
| Synonyms: | AMPA-selective glutamate receptor 1 | GLUH1 | GLUR1 | GRIA1 | GRIA1_HUMAN | GluR-1 | GluR-A | GluR-K1 | Glutamate AMPA 1 | Glutamate receptor 1 | Glutamate receptor AMPA 1/2 | Glutamate receptor ionotropic AMPA | Glutamate receptor ionotropic, AMPA 1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 101512.92 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Glutamate AMPA 1 GRIA1 HUMAN::P42261 |
|---|
| Residue: | 906 |
|---|
| Sequence: | MQHIFAFFCTGFLGAVVGANFPNNIQIGGLFPNQQSQEHAAFRFALSQLTEPPKLLPQID
IVNISDSFEMTYRFCSQFSKGVYAIFGFYERRTVNMLTSFCGALHVCFITPSFPVDTSNQ
FVLQLRPELQDALISIIDHYKWQKFVYIYDADRGLSVLQKVLDTAAEKNWQVTAVNILTT
TEEGYRMLFQDLEKKKERLVVVDCESERLNAILGQIIKLEKNGIGYHYILANLGFMDIDL
NKFKESGANVTGFQLVNYTDTIPAKIMQQWKNSDARDHTRVDWKRPKYTSALTYDGVKVM
AEAFQSLRRQRIDISRRGNAGDCLANPAVPWGQGIDIQRALQQVRFEGLTGNVQFNEKGR
RTNYTLHVIEMKHDGIRKIGYWNEDDKFVPAATDAQAGGDNSSVQNRTYIVTTILEDPYV
MLKKNANQFEGNDRYEGYCVELAAEIAKHVGYSYRLEIVSDGKYGARDPDTKAWNGMVGE
LVYGRADVAVAPLTITLVREEVIDFSKPFMSLGISIMIKKPQKSKPGVFSFLDPLAYEIW
MCIVFAYIGVSVVLFLVSRFSPYEWHSEEFEEGRDQTTSDQSNEFGIFNSLWFSLGAFMQ
QGCDISPRSLSGRIVGGVWWFFTLIIISSYTANLAAFLTVERMVSPIESAEDLAKQTEIA
YGTLEAGSTKEFFRRSKIAVFEKMWTYMKSAEPSVFVRTTEEGMIRVRKSKGKYAYLLES
TMNEYIEQRKPCDTMKVGGNLDSKGYGIATPKGSALRNPVNLAVLKLNEQGLLDKLKNKW
WYDKGECGSGGGDSKDKTSALSLSNVAGVFYILIGGLGLAMLVALIEFCYKSRSESKRMK
GFCLIPQQSINEAIRTSTLPRNSGAGASSGGSGENGRVVSHDFPKSMQSIPCMSHSSGMP
LGATGL
|
|
|
|---|
| BDBM50207594 |
|---|
| n/a |
|---|
| Name | BDBM50207594 |
|---|
| Synonyms: | 2,3-Dihydroxy-6-nitro-benzo[f]quinoxaline-7-sulfonic acid amide | 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo(f)quinoxaline | 2,3-dihydroxy-6-nitro-7-sulfamoylbenzo(f)quinoxaline | 3-Hydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxalin-2-ol anion | 6-Nitro-2,3-dioxo-1,2,3,4,4a,10b-hexahydro-benzo[f]quinoxaline-7-sulfonic acid amide | 6-Nitro-2,3-dioxo-1,2,3,4-tetrahydro-benzo[f]quinoxaline-7-sulfinic acid amide | 6-Nitro-2,3-dioxo-1,2,3,4-tetrahydro-benzo[f]quinoxaline-7-sulfonic acid amide | 6-Nitro-2,3-dioxo-2,3-dihydro-benzo[f]quinoxaline-7-sulfonic acid amide | 6-nitro-2,3-dioxo-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide | CHEMBL222519 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H8N4O6S |
|---|
| Mol. Mass. | 336.28 |
|---|
| SMILES | NS(=O)(=O)c1cccc2c1c(cc1[nH]c(=O)c(=O)[nH]c21)[N+]([O-])=O |
|---|
| Structure | 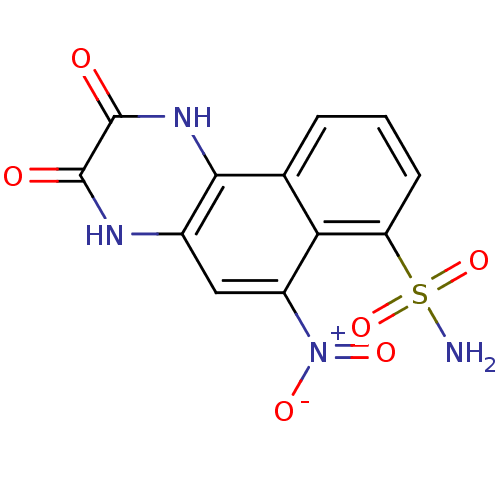
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Filla, SA; Winter, MA; Johnson, KW; Bleakman, D; Bell, MG; Bleisch, TJ; Castaño, AM; Clemens-Smith, A; del Prado, M; Dieckman, DK; Dominguez, E; Escribano, A; Ho, KH; Hudziak, KJ; Katofiasc, MA; Martinez-Perez, JA; Mateo, A; Mathes, BM; Mattiuz, EL; Ogden, AM; Phebus, LA; Stack, DR; Stratford, RE; Ornstein, PL Ethyl (3S,4aR,6S,8aR)-6-(4-ethoxycar- bonylimidazol-1-ylmethyl)decahydroiso-quinoline-3-carboxylic ester: a prodrug of a GluR5 kainate receptor antagonist active in two animal models of acute migraine. J Med Chem45:4383-6 (2002) [PubMed]
Filla, SA; Winter, MA; Johnson, KW; Bleakman, D; Bell, MG; Bleisch, TJ; Castaño, AM; Clemens-Smith, A; del Prado, M; Dieckman, DK; Dominguez, E; Escribano, A; Ho, KH; Hudziak, KJ; Katofiasc, MA; Martinez-Perez, JA; Mateo, A; Mathes, BM; Mattiuz, EL; Ogden, AM; Phebus, LA; Stack, DR; Stratford, RE; Ornstein, PL Ethyl (3S,4aR,6S,8aR)-6-(4-ethoxycar- bonylimidazol-1-ylmethyl)decahydroiso-quinoline-3-carboxylic ester: a prodrug of a GluR5 kainate receptor antagonist active in two animal models of acute migraine. J Med Chem45:4383-6 (2002) [PubMed]