| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 2C9 |
|---|
| Ligand | BDBM50130610 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_51544 (CHEMBL660404) |
|---|
| IC50 | 5300±n/a nM |
|---|
| Citation |  Wu, YJ; Davis, CD; Dworetzky, S; Fitzpatrick, WC; Harden, D; He, H; Knox, RJ; Newton, AE; Philip, T; Polson, C; Sivarao, DV; Sun, LQ; Tertyshnikova, S; Weaver, D; Yeola, S; Zoeckler, M; Sinz, MW Fluorine substitution can block CYP3A4 metabolism-dependent inhibition: identification of (S)-N-[1-(4-fluoro-3- morpholin-4-ylphenyl)ethyl]-3- (4-fluorophenyl)acrylamide as an orally bioavailable KCNQ2 opener devoid of CYP3A4 metabolism-dependent inhibition. J Med Chem46:3778-81 (2003) [PubMed] Article Wu, YJ; Davis, CD; Dworetzky, S; Fitzpatrick, WC; Harden, D; He, H; Knox, RJ; Newton, AE; Philip, T; Polson, C; Sivarao, DV; Sun, LQ; Tertyshnikova, S; Weaver, D; Yeola, S; Zoeckler, M; Sinz, MW Fluorine substitution can block CYP3A4 metabolism-dependent inhibition: identification of (S)-N-[1-(4-fluoro-3- morpholin-4-ylphenyl)ethyl]-3- (4-fluorophenyl)acrylamide as an orally bioavailable KCNQ2 opener devoid of CYP3A4 metabolism-dependent inhibition. J Med Chem46:3778-81 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 2C9 |
|---|
| Name: | Cytochrome P450 2C9 |
|---|
| Synonyms: | (R)-limonene 6-monooxygenase | (S)-limonene 6-monooxygenase | CP2C9_HUMAN | CYP2C10 | CYP2C9 | CYPIIC9 | Cytochrome P450 2C9 (CYP2C9 ) | Cytochrome P450 2C9 (CYP2C9) | P-450MP | P450 MP-4/MP-8 | P450 PB-1 | S-mephenytoin 4-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55636.33 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P11712 |
|---|
| Residue: | 490 |
|---|
| Sequence: | MDSLVVLVLCLSCLLLLSLWRQSSGRGKLPPGPTPLPVIGNILQIGIKDISKSLTNLSKV
YGPVFTLYFGLKPIVVLHGYEAVKEALIDLGEEFSGRGIFPLAERANRGFGIVFSNGKKW
KEIRRFSLMTLRNFGMGKRSIEDRVQEEARCLVEELRKTKASPCDPTFILGCAPCNVICS
IIFHKRFDYKDQQFLNLMEKLNENIKILSSPWIQICNNFSPIIDYFPGTHNKLLKNVAFM
KSYILEKVKEHQESMDMNNPQDFIDCFLMKMEKEKHNQPSEFTIESLENTAVDLFGAGTE
TTSTTLRYALLLLLKHPEVTAKVQEEIERVIGRNRSPCMQDRSHMPYTDAVVHEVQRYID
LLPTSLPHAVTCDIKFRNYLIPKGTTILISLTSVLHDNKEFPNPEMFDPHHFLDEGGNFK
KSKYFMPFSAGKRICVGEALAGMELFLFLTSILQNFNLKSLVDPKNLDTTPVVNGFASVP
PFYQLCFIPV
|
|
|
|---|
| BDBM50130610 |
|---|
| n/a |
|---|
| Name | BDBM50130610 |
|---|
| Synonyms: | (E)-N-[(S)-1-(3-Morpholin-4-yl-phenyl)-ethyl]-3-phenyl-acrylamide | (E)-N-[1-((S)-3-Morpholin-4-yl-phenyl)-ethyl]-3-phenyl-acrylamide | CHEMBL317935 | N-[1-(3-Morpholin-4-yl-phenyl)-ethyl]-3-phenyl-acrylamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H24N2O2 |
|---|
| Mol. Mass. | 336.4275 |
|---|
| SMILES | C[C@H](NC(=O)\C=C\c1ccccc1)c1cccc(c1)N1CCOCC1 |
|---|
| Structure | 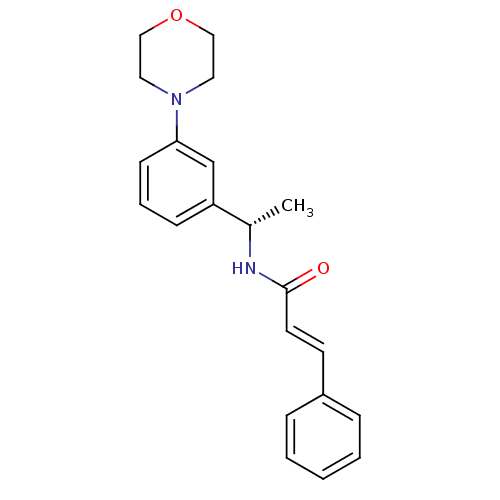
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wu, YJ; Davis, CD; Dworetzky, S; Fitzpatrick, WC; Harden, D; He, H; Knox, RJ; Newton, AE; Philip, T; Polson, C; Sivarao, DV; Sun, LQ; Tertyshnikova, S; Weaver, D; Yeola, S; Zoeckler, M; Sinz, MW Fluorine substitution can block CYP3A4 metabolism-dependent inhibition: identification of (S)-N-[1-(4-fluoro-3- morpholin-4-ylphenyl)ethyl]-3- (4-fluorophenyl)acrylamide as an orally bioavailable KCNQ2 opener devoid of CYP3A4 metabolism-dependent inhibition. J Med Chem46:3778-81 (2003) [PubMed] Article
Wu, YJ; Davis, CD; Dworetzky, S; Fitzpatrick, WC; Harden, D; He, H; Knox, RJ; Newton, AE; Philip, T; Polson, C; Sivarao, DV; Sun, LQ; Tertyshnikova, S; Weaver, D; Yeola, S; Zoeckler, M; Sinz, MW Fluorine substitution can block CYP3A4 metabolism-dependent inhibition: identification of (S)-N-[1-(4-fluoro-3- morpholin-4-ylphenyl)ethyl]-3- (4-fluorophenyl)acrylamide as an orally bioavailable KCNQ2 opener devoid of CYP3A4 metabolism-dependent inhibition. J Med Chem46:3778-81 (2003) [PubMed] Article