| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Prothrombin |
|---|
| Ligand | BDBM50135945 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_208539 (CHEMBL813633) |
|---|
| Ki | 3600±n/a nM |
|---|
| Citation |  Pruitt, JR; Pinto, DJ; Galemmo, RA; Alexander, RS; Rossi, KA; Wells, BL; Drummond, S; Bostrom, LL; Burdick, D; Bruckner, R; Chen, H; Smallwood, A; Wong, PC; Wright, MR; Bai, S; Luettgen, JM; Knabb, RM; Lam, PY; Wexler, RR Discovery of 1-(2-aminomethylphenyl)-3-trifluoromethyl-N- [3-fluoro-2'-(aminosulfonyl)[1,1'-biphenyl)]-4-yl]-1H-pyrazole-5-carboxyamide (DPC602), a potent, selective, and orally bioavailable factor Xa inhibitor(1). J Med Chem46:5298-315 (2003) [PubMed] Article Pruitt, JR; Pinto, DJ; Galemmo, RA; Alexander, RS; Rossi, KA; Wells, BL; Drummond, S; Bostrom, LL; Burdick, D; Bruckner, R; Chen, H; Smallwood, A; Wong, PC; Wright, MR; Bai, S; Luettgen, JM; Knabb, RM; Lam, PY; Wexler, RR Discovery of 1-(2-aminomethylphenyl)-3-trifluoromethyl-N- [3-fluoro-2'-(aminosulfonyl)[1,1'-biphenyl)]-4-yl]-1H-pyrazole-5-carboxyamide (DPC602), a potent, selective, and orally bioavailable factor Xa inhibitor(1). J Med Chem46:5298-315 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Prothrombin |
|---|
| Name: | Prothrombin |
|---|
| Synonyms: | Activation peptide fragment 1 | Activation peptide fragment 2 | Coagulation factor II | F2 | Prothrombin precursor | THRB_HUMAN | Thrombin heavy chain | Thrombin light chain |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 70029.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P00734 |
|---|
| Residue: | 622 |
|---|
| Sequence: | MAHVRGLQLPGCLALAALCSLVHSQHVFLAPQQARSLLQRVRRANTFLEEVRKGNLEREC
VEETCSYEEAFEALESSTATDVFWAKYTACETARTPRDKLAACLEGNCAEGLGTNYRGHV
NITRSGIECQLWRSRYPHKPEINSTTHPGADLQENFCRNPDSSTTGPWCYTTDPTVRRQE
CSIPVCGQDQVTVAMTPRSEGSSVNLSPPLEQCVPDRGQQYQGRLAVTTHGLPCLAWASA
QAKALSKHQDFNSAVQLVENFCRNPDGDEEGVWCYVAGKPGDFGYCDLNYCEEAVEEETG
DGLDEDSDRAIEGRTATSEYQTFFNPRTFGSGEADCGLRPLFEKKSLEDKTERELLESYI
DGRIVEGSDAEIGMSPWQVMLFRKSPQELLCGASLISDRWVLTAAHCLLYPPWDKNFTEN
DLLVRIGKHSRTRYERNIEKISMLEKIYIHPRYNWRENLDRDIALMKLKKPVAFSDYIHP
VCLPDRETAASLLQAGYKGRVTGWGNLKETWTANVGKGQPSVLQVVNLPIVERPVCKDST
RIRITDNMFCAGYKPDEGKRGDACEGDSGGPFVMKSPFNNRWYQMGIVSWGEGCDRDGKY
GFYTHVFRLKKWIQKVIDQFGE
|
|
|
|---|
| BDBM50135945 |
|---|
| n/a |
|---|
| Name | BDBM50135945 |
|---|
| Synonyms: | 1-[2-(aminomethyl)phenyl]-N-(3-fluoro-2'-sulfamoylbiphenyl-4-yl)-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide | 2-(2-Aminomethyl-phenyl)-5-trifluoromethyl-2H-pyrazole-3-carboxylic acid (3-fluoro-2'-sulfamoyl-biphenyl-4-yl)-amide | CHEMBL151939 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H19F4N5O3S |
|---|
| Mol. Mass. | 533.498 |
|---|
| SMILES | NCc1ccccc1-n1nc(cc1C(=O)Nc1ccc(cc1F)-c1ccccc1S(N)(=O)=O)C(F)(F)F |
|---|
| Structure | 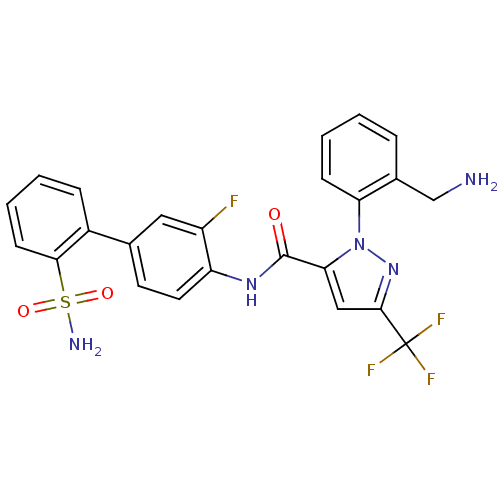
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Pruitt, JR; Pinto, DJ; Galemmo, RA; Alexander, RS; Rossi, KA; Wells, BL; Drummond, S; Bostrom, LL; Burdick, D; Bruckner, R; Chen, H; Smallwood, A; Wong, PC; Wright, MR; Bai, S; Luettgen, JM; Knabb, RM; Lam, PY; Wexler, RR Discovery of 1-(2-aminomethylphenyl)-3-trifluoromethyl-N- [3-fluoro-2'-(aminosulfonyl)[1,1'-biphenyl)]-4-yl]-1H-pyrazole-5-carboxyamide (DPC602), a potent, selective, and orally bioavailable factor Xa inhibitor(1). J Med Chem46:5298-315 (2003) [PubMed] Article
Pruitt, JR; Pinto, DJ; Galemmo, RA; Alexander, RS; Rossi, KA; Wells, BL; Drummond, S; Bostrom, LL; Burdick, D; Bruckner, R; Chen, H; Smallwood, A; Wong, PC; Wright, MR; Bai, S; Luettgen, JM; Knabb, RM; Lam, PY; Wexler, RR Discovery of 1-(2-aminomethylphenyl)-3-trifluoromethyl-N- [3-fluoro-2'-(aminosulfonyl)[1,1'-biphenyl)]-4-yl]-1H-pyrazole-5-carboxyamide (DPC602), a potent, selective, and orally bioavailable factor Xa inhibitor(1). J Med Chem46:5298-315 (2003) [PubMed] Article