| Reaction Details |
|---|
 | Report a problem with these data |
| Target | RNA-directed RNA polymerase |
|---|
| Ligand | BDBM50144936 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_84043 (CHEMBL691889) |
|---|
| IC50 | 50000±n/a nM |
|---|
| Citation |  Eldrup, AB; Allerson, CR; Bennett, CF; Bera, S; Bhat, B; Bhat, N; Bosserman, MR; Brooks, J; Burlein, C; Carroll, SS; Cook, PD; Getty, KL; MacCoss, M; McMasters, DR; Olsen, DB; Prakash, TP; Prhavc, M; Song, Q; Tomassini, JE; Xia, J Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem47:2283-95 (2004) [PubMed] Article Eldrup, AB; Allerson, CR; Bennett, CF; Bera, S; Bhat, B; Bhat, N; Bosserman, MR; Brooks, J; Burlein, C; Carroll, SS; Cook, PD; Getty, KL; MacCoss, M; McMasters, DR; Olsen, DB; Prakash, TP; Prhavc, M; Song, Q; Tomassini, JE; Xia, J Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem47:2283-95 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| RNA-directed RNA polymerase |
|---|
| Name: | RNA-directed RNA polymerase |
|---|
| Synonyms: | Hepatitis C virus NS5B RNA-dependent RNA polymerase | NS5B protein |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 25173.95 |
|---|
| Organism: | Hepatitis C virus |
|---|
| Description: | Q8JXU8 |
|---|
| Residue: | 229 |
|---|
| Sequence: | RTEEAIYQCCDLDPQARVAIRSLTERLYVGGPLTNSRGENCGYRRRASGVLTTSCGNTLT
CYIKAQAACRAAGRQDCTMLVCGDDLVVICESAGVQEDAASLRAFTEAMTRYSAPPGDPP
QPEYDLELITSCSSNVSVAHDGAGKRVYYLTRDPTTPLARAAWETARHTPVNSWLGNIIM
FAPTLWVRMIMLTHFFSVLIARDQLEQALDCEIYGACYSIEPLLPPIIQ
|
|
|
|---|
| BDBM50144936 |
|---|
| n/a |
|---|
| Name | BDBM50144936 |
|---|
| Synonyms: | CHEMBL1090 | VIDARABINE | adenine arabinoside |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H13N5O4 |
|---|
| Mol. Mass. | 267.2413 |
|---|
| SMILES | Nc1ncnc2n(cnc12)[C@@H]1O[C@H](CO)[C@@H](O)[C@@H]1O |r| |
|---|
| Structure | 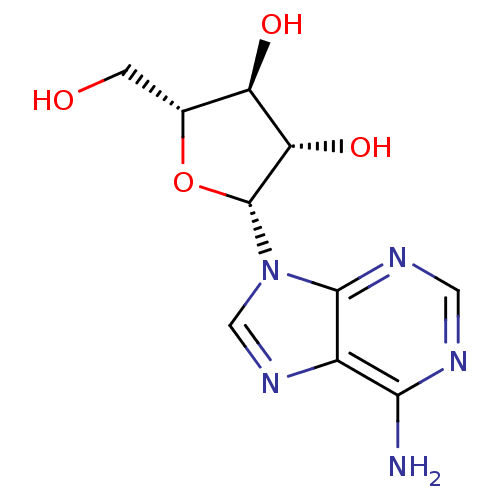
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Eldrup, AB; Allerson, CR; Bennett, CF; Bera, S; Bhat, B; Bhat, N; Bosserman, MR; Brooks, J; Burlein, C; Carroll, SS; Cook, PD; Getty, KL; MacCoss, M; McMasters, DR; Olsen, DB; Prakash, TP; Prhavc, M; Song, Q; Tomassini, JE; Xia, J Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem47:2283-95 (2004) [PubMed] Article
Eldrup, AB; Allerson, CR; Bennett, CF; Bera, S; Bhat, B; Bhat, N; Bosserman, MR; Brooks, J; Burlein, C; Carroll, SS; Cook, PD; Getty, KL; MacCoss, M; McMasters, DR; Olsen, DB; Prakash, TP; Prhavc, M; Song, Q; Tomassini, JE; Xia, J Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem47:2283-95 (2004) [PubMed] Article