

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M1 | ||
| Ligand | BDBM50151215 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_302248 (CHEMBL830264) | ||
| Kd | 19±n/a nM | ||
| Citation |  Tahtaoui, C; Parrot, I; Klotz, P; Guillier, F; Galzi, JL; Hibert, M; Ilien, B Fluorescent pirenzepine derivatives as potential bitopic ligands of the human M1 muscarinic receptor. J Med Chem47:4300-15 (2004) [PubMed] Article Tahtaoui, C; Parrot, I; Klotz, P; Guillier, F; Galzi, JL; Hibert, M; Ilien, B Fluorescent pirenzepine derivatives as potential bitopic ligands of the human M1 muscarinic receptor. J Med Chem47:4300-15 (2004) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M1 | |||
| Name: | Muscarinic acetylcholine receptor M1 | ||
| Synonyms: | ACM1_HUMAN | CHRM1 | Cholinergic receptor, muscarinic 1 | Cholinergic, muscarinic M1 | ||
| Type: | Protein | ||
| Mol. Mass.: | 51442.54 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P11229 | ||
| Residue: | 460 | ||
| Sequence: |
| ||
| BDBM50151215 | |||
| n/a | |||
| Name | BDBM50151215 | ||
| Synonyms: | 2-[6-(diethylamino)-3-(diethyliminiumyl)-3H-xanthen-9-yl]-5-[methyl(5-{[2-(2-{2-[4-(2-oxo-2-{10-oxo-2,4,9-triazatricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,12,14-hexaen-2-yl}ethyl)piperazin-1-yl]ethoxy}ethoxy)ethyl]carbamoyl}pentyl)sulfamoyl]benzene-1-sulfonate | CHEMBL405526 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C58H73N9O11S2 | ||
| Mol. Mass. | 1136.384 | ||
| SMILES | CCN(CC)c1ccc2c(-c3ccc(cc3S([O-])(=O)=O)S(=O)(=O)N(C)CCCCCC(=O)NCCOCCOCCN3CCN(CC(=O)N4c5ccccc5C(=O)Nc5cccnc45)CC3)c3ccc(cc3oc2c1)=[N+](CC)CC |(17.71,10.63,;16.39,9.86,;16.39,8.32,;17.71,7.55,;19.06,8.32,;15.07,7.55,;15.07,6.01,;13.74,5.24,;12.39,6.01,;11.07,5.24,;11.07,3.7,;12.39,2.93,;12.39,1.39,;11.03,.65,;9.71,1.43,;9.71,2.96,;8.22,3.37,;6.9,2.6,;8.22,1.84,;6.9,4.15,;11.03,-.91,;9.49,-.91,;12.55,-.91,;10.24,-2.24,;10.23,-3.77,;8.76,-1.83,;7.4,-2.6,;6.08,-1.83,;4.74,-2.57,;3.4,-1.81,;2.06,-2.57,;2.06,-4.12,;.72,-1.81,;-.62,-1.04,;-1.97,-1.79,;-3.29,-1.03,;-4.62,-1.79,;-5.97,-1,;-7.3,-1.79,;-8.65,-2.56,;-9.97,-1.77,;-11.29,-2.53,;-12.69,-1.79,;-13.98,-2.57,;-13.97,-4.1,;-15.29,-4.91,;-15.26,-6.45,;-14.18,-7.52,;-16.61,-7.26,;-17.89,-6.39,;-17.56,-4.87,;-18.68,-3.84,;-20.14,-4.3,;-20.46,-5.83,;-19.34,-6.86,;-19.89,-8.26,;-21.45,-8.25,;-19.12,-9.61,;-17.6,-9.83,;-17.24,-11.34,;-15.78,-11.77,;-14.67,-10.7,;-15,-9.21,;-16.5,-8.76,;-12.62,-4.87,;-11.29,-4.08,;9.75,6.01,;8.43,5.24,;7.07,6.01,;7.07,7.55,;8.43,8.32,;9.75,7.55,;11.07,8.32,;12.39,7.55,;13.74,8.32,;5.58,7.95,;5.58,9.51,;4.25,10.3,;4.26,7.2,;2.76,7.59,)| | ||
| Structure | 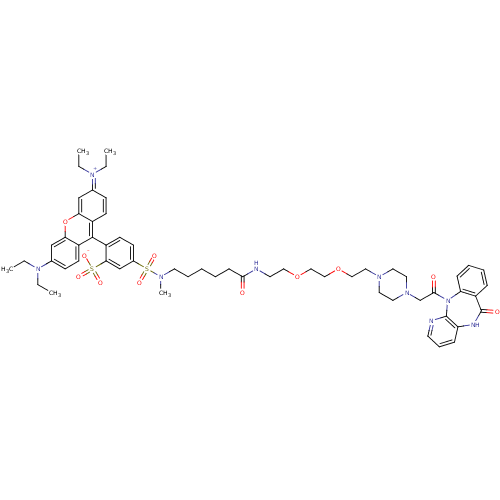
| ||