| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein kinase C epsilon type |
|---|
| Ligand | BDBM50192071 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_404558 (CHEMBL910941) |
|---|
| IC50 | >1000±n/a nM |
|---|
| Citation |  Kawanishi, N; Sugimoto, T; Shibata, J; Nakamura, K; Masutani, K; Ikuta, M; Hirai, H Structure-based drug design of a highly potent CDK1,2,4,6 inhibitor with novel macrocyclic quinoxalin-2-one structure. Bioorg Med Chem Lett16:5122-6 (2006) [PubMed] Article Kawanishi, N; Sugimoto, T; Shibata, J; Nakamura, K; Masutani, K; Ikuta, M; Hirai, H Structure-based drug design of a highly potent CDK1,2,4,6 inhibitor with novel macrocyclic quinoxalin-2-one structure. Bioorg Med Chem Lett16:5122-6 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Protein kinase C epsilon type |
|---|
| Name: | Protein kinase C epsilon type |
|---|
| Synonyms: | KPCE_HUMAN | PKC epsilon | PKCE | PRKCE | Protein kinase C epsilon | Protein kinase C epsilon type | Protein kinase C, PKC; classical/novel | Protein kinase C, epsilon | nPKC-epsilon |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 83680.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | The recombinant human PKC enzymes were
produced using a baculovirus expression system in SF9 cells |
|---|
| Residue: | 737 |
|---|
| Sequence: | MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKT
NSPAWHDEFVTDVCNGRKIELAVFHDAPIGYDDFVANCTIQFEELLQNGSRHFEDWIDLE
PEGRVYVIIDLSGSSGEAPKDNEERVFRERMRPRKRQGAVRRRVHQVNGHKFMATYLRQP
TYCSHCRDFIWGVIGKQGYQCQVCTCVVHKRCHELIITKCAGLKKQETPDQVGSQRFSVN
MPHKFGIHNYKVPTFCDHCGSLLWGLLRQGLQCKVCKMNVHRRCETNVAPNCGVDARGIA
KVLADLGVTPDKITNSGQRRKKLIAGAESPQPASGSSPSEEDRSKSAPTSPCDQEIKELE
NNIRKALSFDNRGEEHRAASSPDGQLMSPGENGEVRQGQAKRLGLDEFNFIKVLGKGSFG
KVMLAELKGKDEVYAVKVLKKDVILQDDDVDCTMTEKRILALARKHPYLTQLYCCFQTKD
RLFFVMEYVNGGDLMFQIQRSRKFDEPRSRFYAAEVTSALMFLHQHGVIYRDLKLDNILL
DAEGHCKLADFGMCKEGILNGVTTTTFCGTPDYIAPEILQELEYGPSVDWWALGVLMYEM
MAGQPPFEADNEDDLFESILHDDVLYPVWLSKEAVSILKAFMTKNPHKRLGCVASQNGED
AIKQHPFFKEIDWVLLEQKKIKPPFKPRIKTKRDVNNFDQDFTREEPVLTLVDEAIVKQI
NQEEFKGFSYFGEDLMP
|
|
|
|---|
| BDBM50192071 |
|---|
| n/a |
|---|
| Name | BDBM50192071 |
|---|
| Synonyms: | (13R,15S)-13-methyl-16-oxa-8,9,12,22,24-pentaazahexacyclo[15.6.2.1^{6,9}.1^{12,15}.0^{2,7}.0^{21,25}]heptacosa-1(24),2,4,6(27),7,17(25),18,20,22-nonaene-23,27-diol | CHEMBL215803 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H21N5O3 |
|---|
| Mol. Mass. | 403.4338 |
|---|
| SMILES | C[C@@H]1C[C@H]2CN1CCn1[nH]c3c(cccc3c1=O)-c1nc3c(O2)cccc3[nH]c1=O |
|---|
| Structure | 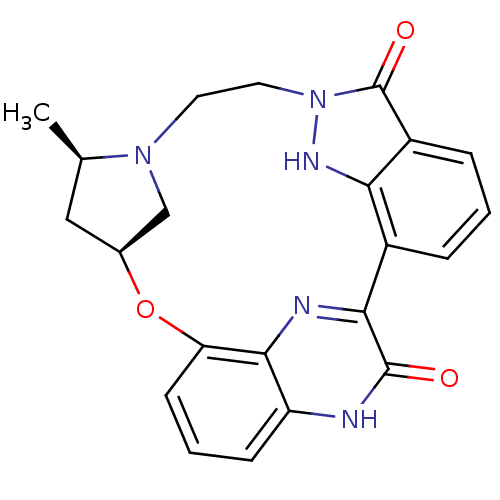
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kawanishi, N; Sugimoto, T; Shibata, J; Nakamura, K; Masutani, K; Ikuta, M; Hirai, H Structure-based drug design of a highly potent CDK1,2,4,6 inhibitor with novel macrocyclic quinoxalin-2-one structure. Bioorg Med Chem Lett16:5122-6 (2006) [PubMed] Article
Kawanishi, N; Sugimoto, T; Shibata, J; Nakamura, K; Masutani, K; Ikuta, M; Hirai, H Structure-based drug design of a highly potent CDK1,2,4,6 inhibitor with novel macrocyclic quinoxalin-2-one structure. Bioorg Med Chem Lett16:5122-6 (2006) [PubMed] Article