| Reaction Details |
|---|
 | Report a problem with these data |
| Target | RNA-directed RNA polymerase |
|---|
| Ligand | BDBM50199056 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_411597 (CHEMBL907248) |
|---|
| IC50 | 77±n/a nM |
|---|
| Citation |  Ikegashira, K; Oka, T; Hirashima, S; Noji, S; Yamanaka, H; Hara, Y; Adachi, T; Tsuruha, J; Doi, S; Hase, Y; Noguchi, T; Ando, I; Ogura, N; Ikeda, S; Hashimoto, H Discovery of conformationally constrained tetracyclic compounds as potent hepatitis C virus NS5B RNA polymerase inhibitors. J Med Chem49:6950-3 (2006) [PubMed] Article Ikegashira, K; Oka, T; Hirashima, S; Noji, S; Yamanaka, H; Hara, Y; Adachi, T; Tsuruha, J; Doi, S; Hase, Y; Noguchi, T; Ando, I; Ogura, N; Ikeda, S; Hashimoto, H Discovery of conformationally constrained tetracyclic compounds as potent hepatitis C virus NS5B RNA polymerase inhibitors. J Med Chem49:6950-3 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| RNA-directed RNA polymerase |
|---|
| Name: | RNA-directed RNA polymerase |
|---|
| Synonyms: | Hepatitis C virus NS5B RNA-dependent RNA polymerase | NS5B protein |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 25173.95 |
|---|
| Organism: | Hepatitis C virus |
|---|
| Description: | Q8JXU8 |
|---|
| Residue: | 229 |
|---|
| Sequence: | RTEEAIYQCCDLDPQARVAIRSLTERLYVGGPLTNSRGENCGYRRRASGVLTTSCGNTLT
CYIKAQAACRAAGRQDCTMLVCGDDLVVICESAGVQEDAASLRAFTEAMTRYSAPPGDPP
QPEYDLELITSCSSNVSVAHDGAGKRVYYLTRDPTTPLARAAWETARHTPVNSWLGNIIM
FAPTLWVRMIMLTHFFSVLIARDQLEQALDCEIYGACYSIEPLLPPIIQ
|
|
|
|---|
| BDBM50199056 |
|---|
| n/a |
|---|
| Name | BDBM50199056 |
|---|
| Synonyms: | 13-cyclohexyl-6,7-dihydro-5H-benzo[3,4]azepino[1,2-a]indole-10-carboxylic acid | CHEMBL216384 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H25NO2 |
|---|
| Mol. Mass. | 359.4608 |
|---|
| SMILES | OC(=O)c1ccc2c(C3CCCCC3)c3-c4ccccc4CCCn3c2c1 |
|---|
| Structure | 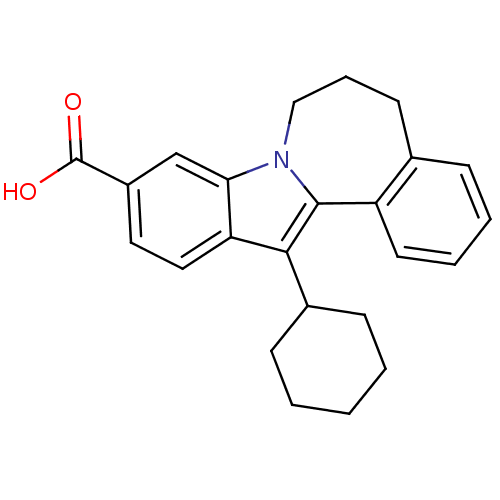
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ikegashira, K; Oka, T; Hirashima, S; Noji, S; Yamanaka, H; Hara, Y; Adachi, T; Tsuruha, J; Doi, S; Hase, Y; Noguchi, T; Ando, I; Ogura, N; Ikeda, S; Hashimoto, H Discovery of conformationally constrained tetracyclic compounds as potent hepatitis C virus NS5B RNA polymerase inhibitors. J Med Chem49:6950-3 (2006) [PubMed] Article
Ikegashira, K; Oka, T; Hirashima, S; Noji, S; Yamanaka, H; Hara, Y; Adachi, T; Tsuruha, J; Doi, S; Hase, Y; Noguchi, T; Ando, I; Ogura, N; Ikeda, S; Hashimoto, H Discovery of conformationally constrained tetracyclic compounds as potent hepatitis C virus NS5B RNA polymerase inhibitors. J Med Chem49:6950-3 (2006) [PubMed] Article