| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM50157136 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_428054 (CHEMBL914372) |
|---|
| Ki | 12±n/a nM |
|---|
| Citation |  Romero, FA; Du, W; Hwang, I; Rayl, TJ; Kimball, FS; Leung, D; Hoover, HS; Apodaca, RL; Breitenbucher, JG; Cravatt, BF; Boger, DL Potent and selective alpha-ketoheterocycle-based inhibitors of the anandamide and oleamide catabolizing enzyme, fatty acid amide hydrolase. J Med Chem50:1058-68 (2007) [PubMed] Article Romero, FA; Du, W; Hwang, I; Rayl, TJ; Kimball, FS; Leung, D; Hoover, HS; Apodaca, RL; Breitenbucher, JG; Cravatt, BF; Boger, DL Potent and selective alpha-ketoheterocycle-based inhibitors of the anandamide and oleamide catabolizing enzyme, fatty acid amide hydrolase. J Med Chem50:1058-68 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM50157136 |
|---|
| n/a |
|---|
| Name | BDBM50157136 |
|---|
| Synonyms: | 1-(5-(furan-2-yl)oxazol-2-yl)-7-phenylheptan-1-one | 1-(5-Furan-2-yl-oxazol-2-yl)-7-phenyl-heptan-1-one | CHEMBL177356 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H21NO3 |
|---|
| Mol. Mass. | 323.3856 |
|---|
| SMILES | O=C(CCCCCCc1ccccc1)c1ncc(o1)-c1ccco1 |
|---|
| Structure | 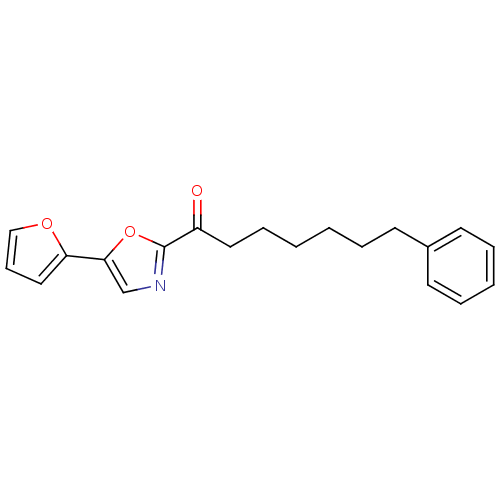
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Romero, FA; Du, W; Hwang, I; Rayl, TJ; Kimball, FS; Leung, D; Hoover, HS; Apodaca, RL; Breitenbucher, JG; Cravatt, BF; Boger, DL Potent and selective alpha-ketoheterocycle-based inhibitors of the anandamide and oleamide catabolizing enzyme, fatty acid amide hydrolase. J Med Chem50:1058-68 (2007) [PubMed] Article
Romero, FA; Du, W; Hwang, I; Rayl, TJ; Kimball, FS; Leung, D; Hoover, HS; Apodaca, RL; Breitenbucher, JG; Cravatt, BF; Boger, DL Potent and selective alpha-ketoheterocycle-based inhibitors of the anandamide and oleamide catabolizing enzyme, fatty acid amide hydrolase. J Med Chem50:1058-68 (2007) [PubMed] Article