| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Vitamin K-dependent protein C |
|---|
| Ligand | BDBM12676 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_461712 (CHEMBL928843) |
|---|
| Ki | >20000±n/a nM |
|---|
| Citation |  Varnes, JG; Wacker, DA; Pinto, DJ; Orwat, MJ; Theroff, JP; Wells, B; Galemo, RA; Luettgen, JM; Knabb, RM; Bai, S; He, K; Lam, PY; Wexler, RR Structure-activity relationship and pharmacokinetic profile of 5-ketopyrazole factor Xa inhibitors. Bioorg Med Chem Lett18:749-54 (2008) [PubMed] Article Varnes, JG; Wacker, DA; Pinto, DJ; Orwat, MJ; Theroff, JP; Wells, B; Galemo, RA; Luettgen, JM; Knabb, RM; Bai, S; He, K; Lam, PY; Wexler, RR Structure-activity relationship and pharmacokinetic profile of 5-ketopyrazole factor Xa inhibitors. Bioorg Med Chem Lett18:749-54 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Vitamin K-dependent protein C |
|---|
| Name: | Vitamin K-dependent protein C |
|---|
| Synonyms: | Activated protein C cofactor | Anticoagulant protein C | Apolipoprotein H | Autoprothrombin IIA | Blood coagulation factor XIV | Coagulation factor V | Coagulation factor V heavy chain | Coagulation factor V light chain | Endothelial protein C receptor | PROC | PROC_HUMAN | Proaccelerin, labile factor | Vitamin K-dependent protein C precursor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52067.73 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 461 |
|---|
| Sequence: | MWQLTSLLLFVATWGISGTPAPLDSVFSSSERAHQVLRIRKRANSFLEELRHSSLERECI
EEICDFEEAKEIFQNVDDTLAFWSKHVDGDQCLVLPLEHPCASLCCGHGTCIDGIGSFSC
DCRSGWEGRFCQREVSFLNCSLDNGGCTHYCLEEVGWRRCSCAPGYKLGDDLLQCHPAVK
FPCGRPWKRMEKKRSHLKRDTEDQEDQVDPRLIDGKMTRRGDSPWQVVLLDSKKKLACGA
VLIHPSWVLTAAHCMDESKKLLVRLGEYDLRRWEKWELDLDIKEVFVHPNYSKSTTDNDI
ALLHLAQPATLSQTIVPICLPDSGLAERELNQAGQETLVTGWGYHSSREKEAKRNRTFVL
NFIKIPVVPHNECSEVMSNMVSENMLCAGILGDRQDACEGDSGGPMVASFHGTWFLVGLV
SWGEGCGLLHNYGVYTKVSRYLDWIHGHIRDKEAPQKSWAP
|
|
|
|---|
| BDBM12676 |
|---|
| n/a |
|---|
| Name | BDBM12676 |
|---|
| Synonyms: | 1-(3-Aminobenzisoxazol-5-yl)-3-trifluoromethyl-N-[2-fluoro-4-[(2-dimethylaminomethyl)imidazol-1-yl]phenyl]-1H-pyrazole-5-carboxyamide Hydrochloride Salt | 1-(3-amino-1,2-benzoxazol-5-yl)-N-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-3-(trifluoromethyl)-1H-pyrazole-5-carboxamide | BMS-561389 | CHEMBL206335 | CHEMBL558198 | DPC 906 | Razaxaban |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H20F4N8O2 |
|---|
| Mol. Mass. | 528.4616 |
|---|
| SMILES | CN(C)Cc1nccn1-c1ccc(NC(=O)c2cc(nn2-c2ccc3onc(N)c3c2)C(F)(F)F)c(F)c1 |
|---|
| Structure | 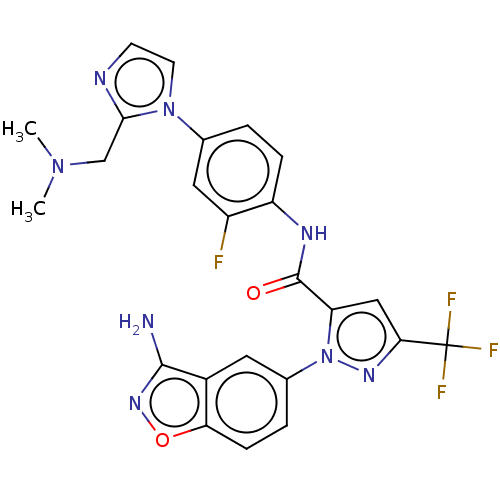
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Varnes, JG; Wacker, DA; Pinto, DJ; Orwat, MJ; Theroff, JP; Wells, B; Galemo, RA; Luettgen, JM; Knabb, RM; Bai, S; He, K; Lam, PY; Wexler, RR Structure-activity relationship and pharmacokinetic profile of 5-ketopyrazole factor Xa inhibitors. Bioorg Med Chem Lett18:749-54 (2008) [PubMed] Article
Varnes, JG; Wacker, DA; Pinto, DJ; Orwat, MJ; Theroff, JP; Wells, B; Galemo, RA; Luettgen, JM; Knabb, RM; Bai, S; He, K; Lam, PY; Wexler, RR Structure-activity relationship and pharmacokinetic profile of 5-ketopyrazole factor Xa inhibitors. Bioorg Med Chem Lett18:749-54 (2008) [PubMed] Article