| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Ligand | BDBM23074 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_464889 (CHEMBL949088) |
|---|
| Ki | 3.4±n/a nM |
|---|
| Citation |  Kimball, FS; Romero, FA; Ezzili, C; Garfunkle, J; Rayl, TJ; Hochstatter, DG; Hwang, I; Boger, DL Optimization of alpha-ketooxazole inhibitors of fatty acid amide hydrolase. J Med Chem51:937-47 (2008) [PubMed] Article Kimball, FS; Romero, FA; Ezzili, C; Garfunkle, J; Rayl, TJ; Hochstatter, DG; Hwang, I; Boger, DL Optimization of alpha-ketooxazole inhibitors of fatty acid amide hydrolase. J Med Chem51:937-47 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Name: | Fatty-acid amide hydrolase 1 [30-579] |
|---|
| Synonyms: | Anandamide amidohydrolase 1 | FAAH1_RAT | Faah | Faah1 | Fatty Acid Amide Hydrolase | Fatty Acid Amide Hydrolic, FAAH | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Fatty-acid amide hydrolase 1 (FAAH) | Fatty-acid amide hydrolase 1 (aa 30-579) | Oleamide hydrolase 1 |
|---|
| Type: | Single-pass membrane protein; homodimer |
|---|
| Mol. Mass.: | 60474.00 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | P97612 (aa 30-579) |
|---|
| Residue: | 550 |
|---|
| Sequence: | RWTGRQKARGAATRARQKQRASLETMDKAVQRFRLQNPDLDSEALLTLPLLQLVQKLQSG
ELSPEAVFFTYLGKAWEVNKGTNCVTSYLTDCETQLSQAPRQGLLYGVPVSLKECFSYKG
HDSTLGLSLNEGMPSESDCVVVQVLKLQGAVPFVHTNVPQSMLSFDCSNPLFGQTMNPWK
SSKSPGGSSGGEGALIGSGGSPLGLGTDIGGSIRFPSAFCGICGLKPTGNRLSKSGLKGC
VYGQTAVQLSLGPMARDVESLALCLKALLCEHLFTLDPTVPPLPFREEVYRSSRPLRVGY
YETDNYTMPSPAMRRALIETKQRLEAAGHTLIPFLPNNIPYALEVLSAGGLFSDGGRSFL
QNFKGDFVDPCLGDLILILRLPSWFKRLLSLLLKPLFPRLAAFLNSMRPRSAEKLWKLQH
EIEMYRQSVIAQWKAMNLDVLLTPMLGPALDLNTPGRATGAISYTVLYNCLDFPAGVVPV
TTVTAEDDAQMELYKGYFGDIWDIILKKAMKNSVGLPVAVQCVALPWQEELCLRFMREVE
QLMTPQKQPS
|
|
|
|---|
| BDBM23074 |
|---|
| n/a |
|---|
| Name | BDBM23074 |
|---|
| Synonyms: | 3-(4-phenoxyphenyl)-1-[5-(pyridin-2-yl)-1,3-oxazol-2-yl]propan-1-one | alpha-ketooxazole, 11f |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H18N2O3 |
|---|
| Mol. Mass. | 370.4006 |
|---|
| SMILES | O=C(CCc1ccc(Oc2ccccc2)cc1)c1ncc(o1)-c1ccccn1 |
|---|
| Structure | 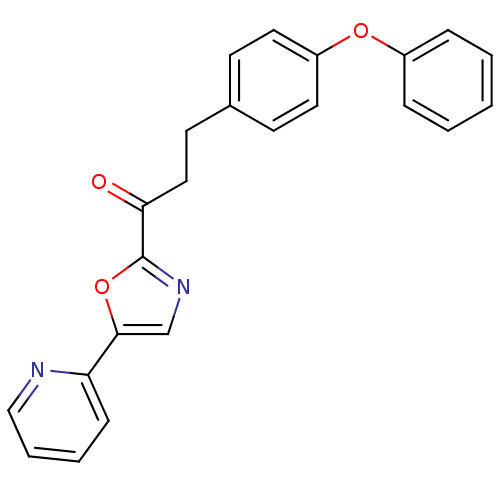
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kimball, FS; Romero, FA; Ezzili, C; Garfunkle, J; Rayl, TJ; Hochstatter, DG; Hwang, I; Boger, DL Optimization of alpha-ketooxazole inhibitors of fatty acid amide hydrolase. J Med Chem51:937-47 (2008) [PubMed] Article
Kimball, FS; Romero, FA; Ezzili, C; Garfunkle, J; Rayl, TJ; Hochstatter, DG; Hwang, I; Boger, DL Optimization of alpha-ketooxazole inhibitors of fatty acid amide hydrolase. J Med Chem51:937-47 (2008) [PubMed] Article