| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 3A4 |
|---|
| Ligand | BDBM50372571 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_465260 (CHEMBL930528) |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Moorjani, M; Zhang, X; Chen, Y; Lin, E; Rueter, JK; Gross, RS; Lanier, MC; Tellew, JE; Williams, JP; Lechner, SM; Malany, S; Santos, M; Ekhlassi, P; Castro-Palomino, JC; Crespo, MI; Prat, M; Gual, S; Díaz, JL; Saunders, J; Slee, DH 2,6-Diaryl-4-phenacylaminopyrimidines as potent and selective adenosine A(2A) antagonists with reduced hERG liability. Bioorg Med Chem Lett18:1269-73 (2008) [PubMed] Article Moorjani, M; Zhang, X; Chen, Y; Lin, E; Rueter, JK; Gross, RS; Lanier, MC; Tellew, JE; Williams, JP; Lechner, SM; Malany, S; Santos, M; Ekhlassi, P; Castro-Palomino, JC; Crespo, MI; Prat, M; Gual, S; Díaz, JL; Saunders, J; Slee, DH 2,6-Diaryl-4-phenacylaminopyrimidines as potent and selective adenosine A(2A) antagonists with reduced hERG liability. Bioorg Med Chem Lett18:1269-73 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 3A4 |
|---|
| Name: | Cytochrome P450 3A4 |
|---|
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 57349.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 503 |
|---|
| Sequence: | MALIPDLAMETWLLLAVSLVLLYLYGTHSHGLFKKLGIPGPTPLPFLGNILSYHKGFCMF
DMECHKKYGKVWGFYDGQQPVLAITDPDMIKTVLVKECYSVFTNRRPFGPVGFMKSAISI
AEDEEWKRLRSLLSPTFTSGKLKEMVPIIAQYGDVLVRNLRREAETGKPVTLKDVFGAYS
MDVITSTSFGVNIDSLNNPQDPFVENTKKLLRFDFLDPFFLSITVFPFLIPILEVLNICV
FPREVTNFLRKSVKRMKESRLEDTQKHRVDFLQLMIDSQNSKETESHKALSDLELVAQSI
IFIFAGYETTSSVLSFIMYELATHPDVQQKLQEEIDAVLPNKAPPTYDTVLQMEYLDMVV
NETLRLFPIAMRLERVCKKDVEINGMFIPKGVVVMIPSYALHRDPKYWTEPEKFLPERFS
KKNKDNIDPYIYTPFGSGPRNCIGMRFALMNMKLALIRVLQNFSFKPCKETQIPLKLSLG
GLLQPEKPVVLKVESRDGTVSGA
|
|
|
|---|
| BDBM50372571 |
|---|
| n/a |
|---|
| Name | BDBM50372571 |
|---|
| Synonyms: | CHEMBL270654 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H22N6O3S |
|---|
| Mol. Mass. | 462.524 |
|---|
| SMILES | Cc1cc(C)n(n1)-c1cc(NC(=O)Cc2ccc(cc2)S(C)(=O)=O)nc(n1)-c1ccccn1 |
|---|
| Structure | 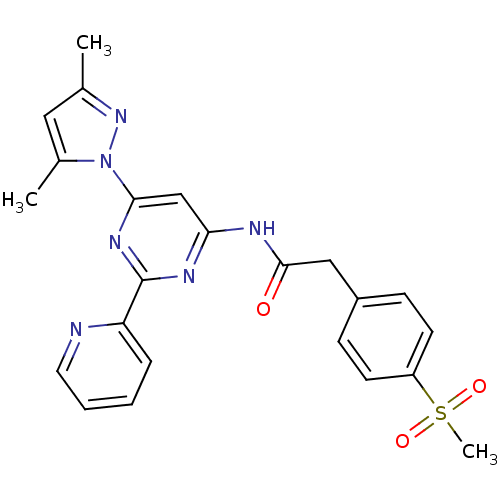
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Moorjani, M; Zhang, X; Chen, Y; Lin, E; Rueter, JK; Gross, RS; Lanier, MC; Tellew, JE; Williams, JP; Lechner, SM; Malany, S; Santos, M; Ekhlassi, P; Castro-Palomino, JC; Crespo, MI; Prat, M; Gual, S; Díaz, JL; Saunders, J; Slee, DH 2,6-Diaryl-4-phenacylaminopyrimidines as potent and selective adenosine A(2A) antagonists with reduced hERG liability. Bioorg Med Chem Lett18:1269-73 (2008) [PubMed] Article
Moorjani, M; Zhang, X; Chen, Y; Lin, E; Rueter, JK; Gross, RS; Lanier, MC; Tellew, JE; Williams, JP; Lechner, SM; Malany, S; Santos, M; Ekhlassi, P; Castro-Palomino, JC; Crespo, MI; Prat, M; Gual, S; Díaz, JL; Saunders, J; Slee, DH 2,6-Diaryl-4-phenacylaminopyrimidines as potent and selective adenosine A(2A) antagonists with reduced hERG liability. Bioorg Med Chem Lett18:1269-73 (2008) [PubMed] Article