| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 1 |
|---|
| Ligand | BDBM50229978 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_473589 (CHEMBL939354) |
|---|
| Ki | 310±n/a nM |
|---|
| Citation |  Fedorov, O; Marsden, B; Pogacic, V; Rellos, P; Müller, S; Bullock, AN; Schwaller, J; Sundström, M; Knapp, S A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A104:20523-8 (2007) [PubMed] Article Fedorov, O; Marsden, B; Pogacic, V; Rellos, P; Müller, S; Bullock, AN; Schwaller, J; Sundström, M; Knapp, S A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A104:20523-8 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 1 |
|---|
| Name: | Mitogen-activated protein kinase 1 |
|---|
| Synonyms: | ERK2 | ERT1 | Extracellular signal-regulated kinase 2 | Extracellular signal-regulated kinase 2 (ERK-2) | Extracellular signal-regulated kinase 2 (ERK2) | MAP Kinase 2/ERK2 | MAPK 2 | MAPK1 | MK01_HUMAN | Mitogen activated kinase 1 (ERK-2) | Mitogen-activated protein kinase 1 (ERK-2) | Mitogen-activated protein kinase 1 (ERK2) | Mitogen-activated protein kinase 2 | PRKM1 | PRKM2 | p42-MAPK |
|---|
| Type: | Ser/Thr Protein Kinase |
|---|
| Mol. Mass.: | 41392.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P28482 |
|---|
| Residue: | 360 |
|---|
| Sequence: | MAAAAAAGAGPEMVRGQVFDVGPRYTNLSYIGEGAYGMVCSAYDNVNKVRVAIKKISPFE
HQTYCQRTLREIKILLRFRHENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQH
LSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTTCDLKICDFGLARVADPDHDH
TGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLSNRPIFPGKHYLDQLNHI
LGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSKALDLLDKMLTFNPHK
RIEVEQALAHPYLEQYYDPSDEPIAEAPFKFDMELDDLPKEKLKELIFEETARFQPGYRS
|
|
|
|---|
| BDBM50229978 |
|---|
| n/a |
|---|
| Name | BDBM50229978 |
|---|
| Synonyms: | 5-(2-PHENYLPYRAZOLO[1,5-A]PYRIDIN-3-YL)-1H-PYRAZOLO[3,4-C]PYRIDAZIN-3-AMINE | 5-(2-Phenyl-pyrazolo[1,5-a]pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridazin-3 -ylamine | 5-(2-phenylH-pyrazolo[1,5-a]pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridazin-3-amine | CHEMBL259551 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H13N7 |
|---|
| Mol. Mass. | 327.3427 |
|---|
| SMILES | Nc1n[nH]c2nnc(cc12)-c1c(nn2ccccc12)-c1ccccc1 |
|---|
| Structure | 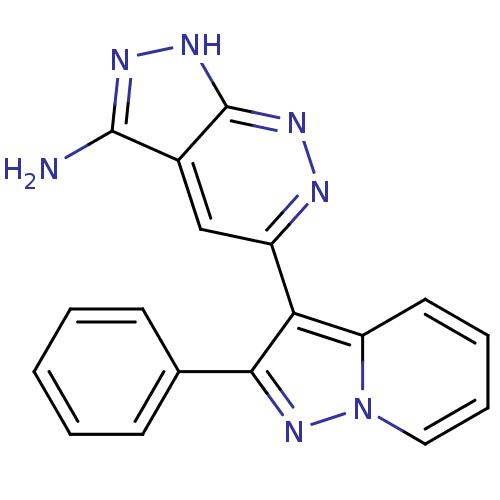
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Fedorov, O; Marsden, B; Pogacic, V; Rellos, P; Müller, S; Bullock, AN; Schwaller, J; Sundström, M; Knapp, S A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A104:20523-8 (2007) [PubMed] Article
Fedorov, O; Marsden, B; Pogacic, V; Rellos, P; Müller, S; Bullock, AN; Schwaller, J; Sundström, M; Knapp, S A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A104:20523-8 (2007) [PubMed] Article