| Reaction Details |
|---|
 | Report a problem with these data |
| Target | RAC-beta serine/threonine-protein kinase |
|---|
| Ligand | BDBM50237622 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_475113 (CHEMBL932635) |
|---|
| IC50 | 6±n/a nM |
|---|
| Citation |  Caldwell, JJ; Davies, TG; Donald, A; McHardy, T; Rowlands, MG; Aherne, GW; Hunter, LK; Taylor, K; Ruddle, R; Raynaud, FI; Verdonk, M; Workman, P; Garrett, MD; Collins, I Identification of 4-(4-aminopiperidin-1-yl)-7H-pyrrolo[2,3-d]pyrimidines as selective inhibitors of protein kinase B through fragment elaboration. J Med Chem51:2147-57 (2008) [PubMed] Article Caldwell, JJ; Davies, TG; Donald, A; McHardy, T; Rowlands, MG; Aherne, GW; Hunter, LK; Taylor, K; Ruddle, R; Raynaud, FI; Verdonk, M; Workman, P; Garrett, MD; Collins, I Identification of 4-(4-aminopiperidin-1-yl)-7H-pyrrolo[2,3-d]pyrimidines as selective inhibitors of protein kinase B through fragment elaboration. J Med Chem51:2147-57 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| RAC-beta serine/threonine-protein kinase |
|---|
| Name: | RAC-beta serine/threonine-protein kinase |
|---|
| Synonyms: | AKT2 | AKT2_HUMAN | PKB beta | Protein kinase Akt-2 | Protein kinase B (Akt 2) | Protein kinase B beta (AKT2) | Protein kinase B, beta | RAC-PK-beta | RAC-alpha serine/threonine-protein kinase (AKT2) | RAC-beta serine/threonine-protein kinase | RAC-beta serine/threonine-protein kinase (AKT2) | Serine/threonine-protein kinase AKT | Serine/threonine-protein kinase AKT2 | Serine/threonine-protein kinase Rac alpha/beta |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55766.64 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 481 |
|---|
| Sequence: | MNEVSVIKEGWLHKRGEYIKTWRPRYFLLKSDGSFIGYKERPEAPDQTLPPLNNFSVAEC
QLMKTERPRPNTFVIRCLQWTTVIERTFHVDSPDEREEWMRAIQMVANSLKQRAPGEDPM
DYKCGSPSDSSTTEEMEVAVSKARAKVTMNDFDYLKLLGKGTFGKVILVREKATGRYYAM
KILRKEVIIAKDEVAHTVTESRVLQNTRHPFLTALKYAFQTHDRLCFVMEYANGGELFFH
LSRERVFTEERARFYGAEIVSALEYLHSRDVVYRDIKLENLMLDKDGHIKITDFGLCKEG
ISDGATMKTFCGTPEYLAPEVLEDNDYGRAVDWWGLGVVMYEMMCGRLPFYNQDHERLFE
LILMEEIRFPRTLSPEAKSLLAGLLKKDPKQRLGGGPSDAKEVMEHRFFLSINWQDVVQK
KLLPPFKPQVTSEVDTRYFDDEFTAQSITITPPDRYDSLGLLELDQRTHFPQFSYSASIR
E
|
|
|
|---|
| BDBM50237622 |
|---|
| n/a |
|---|
| Name | BDBM50237622 |
|---|
| Synonyms: | 4-(4-Chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin- | 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-amine | CCT128930 | CHEMBL263664 | US8796293, 17 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H20ClN5 |
|---|
| Mol. Mass. | 341.838 |
|---|
| SMILES | NC1(Cc2ccc(Cl)cc2)CCN(CC1)c1ncnc2[nH]ccc12 |
|---|
| Structure | 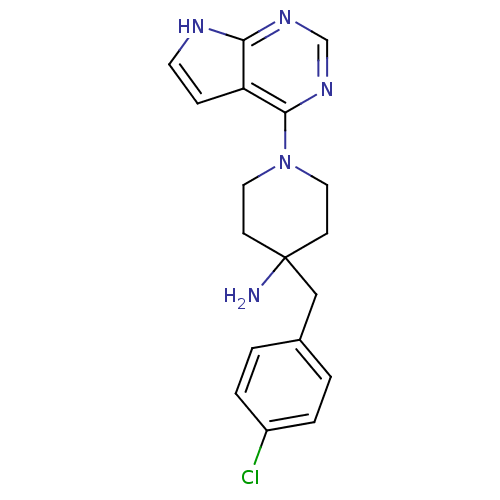
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Caldwell, JJ; Davies, TG; Donald, A; McHardy, T; Rowlands, MG; Aherne, GW; Hunter, LK; Taylor, K; Ruddle, R; Raynaud, FI; Verdonk, M; Workman, P; Garrett, MD; Collins, I Identification of 4-(4-aminopiperidin-1-yl)-7H-pyrrolo[2,3-d]pyrimidines as selective inhibitors of protein kinase B through fragment elaboration. J Med Chem51:2147-57 (2008) [PubMed] Article
Caldwell, JJ; Davies, TG; Donald, A; McHardy, T; Rowlands, MG; Aherne, GW; Hunter, LK; Taylor, K; Ruddle, R; Raynaud, FI; Verdonk, M; Workman, P; Garrett, MD; Collins, I Identification of 4-(4-aminopiperidin-1-yl)-7H-pyrrolo[2,3-d]pyrimidines as selective inhibitors of protein kinase B through fragment elaboration. J Med Chem51:2147-57 (2008) [PubMed] Article