| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dipeptidyl peptidase 2 |
|---|
| Ligand | BDBM50050525 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_541739 (CHEMBL1023101) |
|---|
| Ki | 125±n/a nM |
|---|
| Citation |  Havale, SH; Pal, M Medicinal chemistry approaches to the inhibition of dipeptidyl peptidase-4 for the treatment of type 2 diabetes. Bioorg Med Chem17:1783-802 (2009) [PubMed] Article Havale, SH; Pal, M Medicinal chemistry approaches to the inhibition of dipeptidyl peptidase-4 for the treatment of type 2 diabetes. Bioorg Med Chem17:1783-802 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dipeptidyl peptidase 2 |
|---|
| Name: | Dipeptidyl peptidase 2 |
|---|
| Synonyms: | DAP II | DPP2 | DPP2_HUMAN | DPP7 | Dipeptidyl aminopeptidase II | Dipeptidyl peptidase 2 (DPP II) | Dipeptidyl peptidase 2 (DPP2) | Dipeptidyl peptidase II (DDP-II) | Dipeptidyl peptidase II (DPP II) | Dipeptidyl peptidase II (DPP2) | Dipeptidyl peptidase II and dipeptidyl peptidase IV (DPP2 and DPP4) | QPP | carboxytripeptidase | dipeptidyl arylamidase II | dipeptidyl(amino)peptidase II | dipeptidylarylamidase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 54339.29 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UHL4 |
|---|
| Residue: | 492 |
|---|
| Sequence: | MGSAPWAPVLLLALGLRGLQAGARRAPDPGFQERFFQQRLDHFNFERFGNKTFPQRFLVS
DRFWVRGEGPIFFYTGNEGDVWAFANNSAFVAELAAERGALLVFAEHRYYGKSLPFGAQS
TQRGHTELLTVEQALADFAELLRALRRDLGAQDAPAIAFGGSYGGMLSAYLRMKYPHLVA
GALAASAPVLAVAGLGDSNQFFRDVTADFEGQSPKCTQGVREAFRQIKDLFLQGAYDTVR
WEFGTCQPLSDEKDLTQLFMFARNAFTVLAMMDYPYPTDFLGPLPANPVKVGCDRLLSEA
QRITGLRALAGLVYNASGSEHCYDIYRLYHSCADPTGCGTGPDARAWDYQACTEINLTFA
SNNVTDMFPDLPFTDELRQRYCLDTWGVWPRPDWLLTSFWGGDLRAASNIIFSNGNLDPW
AGGGIRRNLSASVIAVTIQGGAHHLDLRASHPEDPASVVEARKLEATIIGEWVKAARREQ
QPALRGGPRLSL
|
|
|
|---|
| BDBM50050525 |
|---|
| n/a |
|---|
| Name | BDBM50050525 |
|---|
| Synonyms: | (R)-1-((S)-2-aminopropanoyl)pyrrolidin-2-ylboronic acid | Boronic acid derivative | CHEMBL63860 | US11096924, DASH-inhibitors 1129 | US11559537, Compound 1129 | US8933056, Ala-boroPro | US8933056, Asp-boroPro |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C7H15BN2O3 |
|---|
| Mol. Mass. | 186.017 |
|---|
| SMILES | C[C@H](N)C(=O)N1CCC[C@H]1B(O)O |r| |
|---|
| Structure | 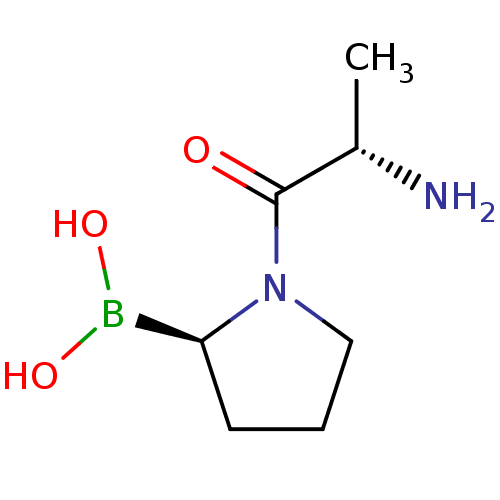
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Havale, SH; Pal, M Medicinal chemistry approaches to the inhibition of dipeptidyl peptidase-4 for the treatment of type 2 diabetes. Bioorg Med Chem17:1783-802 (2009) [PubMed] Article
Havale, SH; Pal, M Medicinal chemistry approaches to the inhibition of dipeptidyl peptidase-4 for the treatment of type 2 diabetes. Bioorg Med Chem17:1783-802 (2009) [PubMed] Article