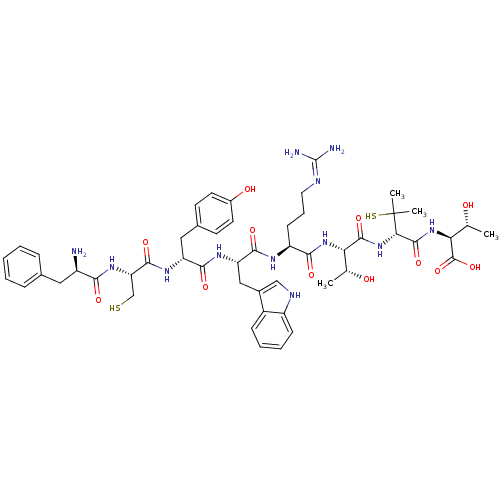
| SMILES | C[C@@H](O)[C@H](NC(=O)[C@H](NC(=O)[C@@H](NC(=O)[C@H](CCCN=C(N)N)NC(=O)[C@H](Cc1c[nH]c2ccccc12)NC(=O)[C@@H](Cc1ccc(O)cc1)NC(=O)[C@H](CS)NC(=O)[C@H](N)Cc1ccccc1)[C@@H](C)O)C(C)(C)S)C(O)=O |r,wU:40.51,26.38,11.11,67.72,wD:7.7,3.3,58.61,52.57,15.22,1.1,(40.38,-16.39,;41.72,-15.62,;43.06,-16.39,;41.72,-14.08,;40.39,-13.31,;39.06,-14.08,;39.06,-15.62,;37.72,-13.31,;36.39,-14.08,;36.39,-15.62,;37.72,-16.39,;35.06,-16.39,;35.06,-17.93,;33.72,-18.7,;32.39,-17.92,;33.72,-20.24,;35.06,-21.01,;36.39,-20.24,;37.72,-21,;39.05,-20.23,;40.39,-21,;40.39,-22.54,;41.72,-20.23,;32.39,-21.01,;32.39,-22.55,;33.72,-23.31,;31.06,-23.31,;29.72,-22.54,;29.72,-21.01,;30.96,-20.1,;30.48,-18.64,;28.94,-18.64,;27.91,-17.49,;26.4,-17.82,;25.93,-19.28,;26.96,-20.42,;28.47,-20.1,;31.06,-24.85,;29.72,-25.62,;28.39,-24.85,;29.72,-27.17,;31.06,-27.94,;32.39,-27.16,;33.73,-27.93,;35.06,-27.16,;35.06,-25.62,;36.39,-24.85,;33.72,-24.85,;32.39,-25.62,;28.39,-27.94,;27.06,-27.17,;27.06,-25.63,;25.72,-27.94,;25.72,-29.48,;27.06,-30.25,;24.39,-27.17,;23.05,-27.94,;23.05,-29.48,;21.72,-27.17,;20.38,-27.94,;21.72,-25.63,;23.05,-24.86,;24.38,-25.63,;25.72,-24.86,;25.72,-23.32,;24.38,-22.55,;23.05,-23.32,;33.72,-15.61,;32.38,-16.39,;33.72,-14.08,;37.72,-11.77,;39.26,-11.76,;36.18,-11.78,;37.71,-10.22,;43.06,-13.3,;44.4,-14.07,;43.06,-11.74,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, G; Aschenbach, LC; He, H; Selley, DE; Zhang, Y 14-O-Heterocyclic-substituted naltrexone derivatives as non-peptide mu opioid receptor selective antagonists: design, synthesis, and biological studies. Bioorg Med Chem Lett19:1825-9 (2009) [PubMed] Article
Li, G; Aschenbach, LC; He, H; Selley, DE; Zhang, Y 14-O-Heterocyclic-substituted naltrexone derivatives as non-peptide mu opioid receptor selective antagonists: design, synthesis, and biological studies. Bioorg Med Chem Lett19:1825-9 (2009) [PubMed] Article