| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histamine H3 receptor |
|---|
| Ligand | BDBM50193199 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_523447 (CHEMBL1008299) |
|---|
| Ki | 7±n/a nM |
|---|
| Citation |  Pierson, PD; Fettes, A; Freichel, C; Gatti-McArthur, S; Hertel, C; Huwyler, J; Mohr, P; Nakagawa, T; Nettekoven, M; Plancher, JM; Raab, S; Richter, H; Roche, O; Rodríguez Sarmiento, RM; Schmitt, M; Schuler, F; Takahashi, T; Taylor, S; Ullmer, C; Wiegand, R 5-hydroxyindole-2-carboxylic acid amides: novel histamine-3 receptor inverse agonists for the treatment of obesity. J Med Chem52:3855-68 (2009) [PubMed] Article Pierson, PD; Fettes, A; Freichel, C; Gatti-McArthur, S; Hertel, C; Huwyler, J; Mohr, P; Nakagawa, T; Nettekoven, M; Plancher, JM; Raab, S; Richter, H; Roche, O; Rodríguez Sarmiento, RM; Schmitt, M; Schuler, F; Takahashi, T; Taylor, S; Ullmer, C; Wiegand, R 5-hydroxyindole-2-carboxylic acid amides: novel histamine-3 receptor inverse agonists for the treatment of obesity. J Med Chem52:3855-68 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histamine H3 receptor |
|---|
| Name: | Histamine H3 receptor |
|---|
| Synonyms: | G-protein coupled receptor 97 | GPCR97 | HH3R | HISTAMINE H3 | HRH3 | HRH3_HUMAN | Histamine H3 receptor (H3) | Histamine H3L | Histamine receptor (H3 and H4) |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 48691.47 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Binding assays were using CHO cells stably expressing hH3R receptors. |
|---|
| Residue: | 445 |
|---|
| Sequence: | MERAPPDGPLNASGALAGEAAAAGGARGFSAAWTAVLAALMALLIVATVLGNALVMLAFV
ADSSLRTQNNFFLLNLAISDFLVGAFCIPLYVPYVLTGRWTFGRGLCKLWLVVDYLLCTS
SAFNIVLISYDRFLSVTRAVSYRAQQGDTRRAVRKMLLVWVLAFLLYGPAILSWEYLSGG
SSIPEGHCYAEFFYNWYFLITASTLEFFTPFLSVTFFNLSIYLNIQRRTRLRLDGAREAA
GPEPPPEAQPSPPPPPGCWGCWQKGHGEAMPLHRYGVGEAAVGAEAGEATLGGGGGGGSV
ASPTSSSGSSSRGTERPRSLKRGSKPSASSASLEKRMKMVSQSFTQRFRLSRDRKVAKSL
AVIVSIFGLCWAPYTLLMIIRAACHGHCVPDYWYETSFWLLWANSAVNPVLYPLCHHSFR
RAFTKLLCPQKLKIQPHSSLEHCWK
|
|
|
|---|
| BDBM50193199 |
|---|
| n/a |
|---|
| Name | BDBM50193199 |
|---|
| Synonyms: | (S)-1-(2-(pyrrolidin-1-ylmethyl)pyrrolidin-1-yl)-3-(4-(trifluoromethyl)phenyl)prop-2-en-1-one | (S,E)-1-(2-(pyrrolidin-1-ylmethyl)pyrrolidin-1-yl)-3-(4-(trifluoromethyl)phenyl)prop-2-en-1-one | CHEMBL218790 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H23F3N2O |
|---|
| Mol. Mass. | 352.3939 |
|---|
| SMILES | FC(F)(F)c1ccc(\C=C\C(=O)N2CCC[C@H]2CN2CCCC2)cc1 |r| |
|---|
| Structure | 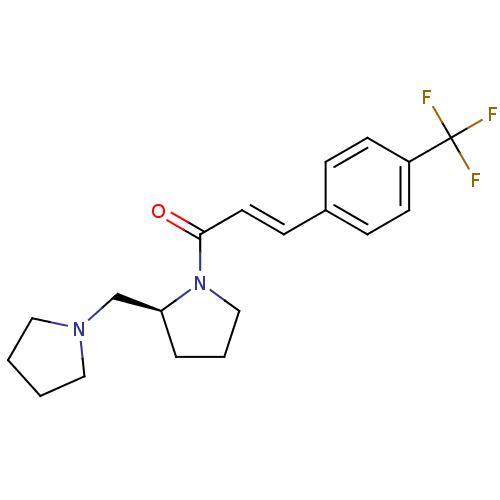
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Pierson, PD; Fettes, A; Freichel, C; Gatti-McArthur, S; Hertel, C; Huwyler, J; Mohr, P; Nakagawa, T; Nettekoven, M; Plancher, JM; Raab, S; Richter, H; Roche, O; Rodríguez Sarmiento, RM; Schmitt, M; Schuler, F; Takahashi, T; Taylor, S; Ullmer, C; Wiegand, R 5-hydroxyindole-2-carboxylic acid amides: novel histamine-3 receptor inverse agonists for the treatment of obesity. J Med Chem52:3855-68 (2009) [PubMed] Article
Pierson, PD; Fettes, A; Freichel, C; Gatti-McArthur, S; Hertel, C; Huwyler, J; Mohr, P; Nakagawa, T; Nettekoven, M; Plancher, JM; Raab, S; Richter, H; Roche, O; Rodríguez Sarmiento, RM; Schmitt, M; Schuler, F; Takahashi, T; Taylor, S; Ullmer, C; Wiegand, R 5-hydroxyindole-2-carboxylic acid amides: novel histamine-3 receptor inverse agonists for the treatment of obesity. J Med Chem52:3855-68 (2009) [PubMed] Article