| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Myosin light chain kinase, smooth muscle |
|---|
| Ligand | BDBM50366348 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_587164 (CHEMBL1060228) |
|---|
| Ki | 97000±n/a nM |
|---|
| Citation |  Engh, RA; Girod, A; Kinzel, V; Huber, R; Bossemeyer, D Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity. J Biol Chem271:26157-64 (1996) [PubMed] Engh, RA; Girod, A; Kinzel, V; Huber, R; Bossemeyer, D Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity. J Biol Chem271:26157-64 (1996) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Myosin light chain kinase, smooth muscle |
|---|
| Name: | Myosin light chain kinase, smooth muscle |
|---|
| Synonyms: | MLCK | MYLK_CHICK | Mylk | Myosin Light-Chain Kinase | Telokin |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 210423.57 |
|---|
| Organism: | Gallus gallus (chicken) |
|---|
| Description: | n/a |
|---|
| Residue: | 1906 |
|---|
| Sequence: | MGDVKLVTSTRVSKTSLTLSPSVPAEAPAFTLPPRNIRVQLGATARFEGKVRGYPEPQIT
WYRNGHPLPEGDHYVVDHSIRGIFSLVIKGVQEGDSGKYTCEAANDGGVRQVTVELTVEG
NSLKKYSLPSSAKTPGGRLSVPPVEHRPSIWGESPPKFATKPNRVVVREGQTGRFSCKIT
GRPQPQVTWTKGDIHLQQNERFNMFEKTGIQYLEIQNVQLADAGIYTCTVVNSAGKASVS
AELTVQGPDKTDTHAQPLCMPPKPTTLATKAIENSDFKQATSNGIAKELKSTSTELMVET
KDRLSAKKETFYTSREAKDGKQGQNQEANAVPLQESRGTKGPQVLQKTSSTITLQAVKAQ
PEPKAEPQTTFIRQAEDRKRTVQPLMTTTTQENPSLTGQVSPRSRETENRAGVRKSVKEE
KREPLGIPPQFESRPQSLEASEGQEIKFKSKVSGKPKPDVEWFKEGVPIKTGEGIQIYEE
DGTHCLWLKKACLGDSGSYSCAAFNPRGQTSTSWLLTVKRPKVEEVAPCFSSVLKGCTVS
EGQDFVLQCYVGGVPVPEITWLLNEQPIQYAHSTFEAGVAKLTVQDALPEDDGIYTCLAE
NNAGRASCSAQVTVKEKKSSKKAEGTQAAKLNKTFAPIFLKGLTDLKVMDGSQVIMTVEV
SANPCPEIIWLHNGKEIQETEDFHFEKKGNEYSLYIQEVFPEDTGKYTCEAWNELGETQT
QATLTVQEPQDGIQPWFISKPRSVTAAAGQNVLISCAIAGDPFPTVHWFKDGQEITPGTG
CEILQNEDIFTLILRNVQSRHAGQYEIQLRNQVGECSCQVSLMLRESSASRAEMLRDGRE
SASSGERRDGGNYGALTFGRTSGFKKSSSETRAAEEEQEDVRGVLKRRVETREHTEESLR
QQEAEQLDFRDILGKKVSTKSFSEEDLKEIPAEQMDFRANLQRQVKPKTLSEEERKVHAP
QQVDFRSVLAKKGTPKTPLPEKVPPPKPAVTDFRSVLGAKKKPPAENGSASTPAPNARAG
SEAQNATPNSEAPAPKPVVKKEEKNDRKCEHGCAVVDGGIIGKKAENKPAASKPTPPPSK
GTAPSFTEKLQDAKVADGEKLVLQCRISSDPPASVSWTLDSKAIKSSKSIVISQEGTLCS
LTIEKVMPEDGGEYKCIAENAAGKAECACKVLVEDTSSTKAAKPAEKKTKKPKTTLPPVL
STESSEATVKKKPAPKTPPKAATPPQITQFPEDRKVRAGESVELFAKVVGTAPITCTWMK
FRKQIQENEYIKIENAENSSKLTISSTKQEHCGCYTLVVENKLGSRQAQVNLTVVDKPDP
PAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYTVEIWNSVDNKWTDLTTCRSTSFNVQD
LQADREYKFRVRAANVYGISEPSQESEVVKVGEKQEEELKEEEAELSDDEGKETEVNYRT
VTINTEQKVSDVYNIEERLGSGKFGQVFRLVEKKTGKVWAGKFFKAYSAKEKENIRDEIS
IMNCLHHPKLVQCVDAFEEKANIVMVLEMVSGGELFERIIDEDFELTERECIKYMRQISE
GVEYIHKQGIVHLDLKPENIMCVNKTGTSIKLIDFGLARRLESAGSLKVLFGTPEFVAPE
VINYEPIGYETDMWSIGVICYILVSGLSPFMGDNDNETLANVTSATWDFDDEAFDEISDD
AKDFISNLLKKDMKSRLNCTQCLQHPWLQKDTKNMEAKKLSKDRMKKYMARRKWQKTGHA
VRAIGRLSSMAMISGMSGRKASGSSPTSPINADKVENEDAFLEEVAEEKPHVKPYFTKTI
LDMEVVEGSAARFDCKIEGYPDPEVMWYKDDQPVKESRHFQIDYDEEGNCSLTISEVCGD
DDAKYTCKAVNSLGEATCTAELLVETMGKEGEGEGEGEEDEEEEEE
|
|
|
|---|
| BDBM50366348 |
|---|
| n/a |
|---|
| Name | BDBM50366348 |
|---|
| Synonyms: | CHEMBL1233654 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H17N3O2S |
|---|
| Mol. Mass. | 291.369 |
|---|
| SMILES | C[C@H]1CNCCN1S(=O)(=O)c1cccc2cnccc12 |r| |
|---|
| Structure | 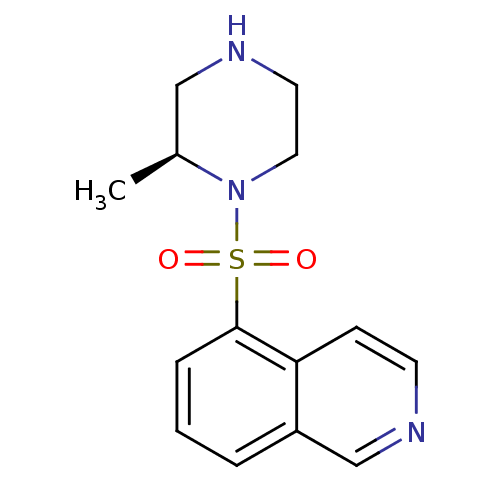
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Engh, RA; Girod, A; Kinzel, V; Huber, R; Bossemeyer, D Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity. J Biol Chem271:26157-64 (1996) [PubMed]
Engh, RA; Girod, A; Kinzel, V; Huber, R; Bossemeyer, D Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity. J Biol Chem271:26157-64 (1996) [PubMed]