

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cannabinoid receptor 2 | ||
| Ligand | BDBM50298951 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_588142 (CHEMBL1049259) | ||
| Ki | >3940±n/a nM | ||
| Citation |  Silvestri, R; Ligresti, A; La Regina, G; Piscitelli, F; Gatti, V; Brizzi, A; Pasquini, S; Lavecchia, A; Allarà, M; Fantini, N; Carai, MA; Novellino, E; Colombo, G; Di Marzo, V; Corelli, F Synthesis, cannabinoid receptor affinity, molecular modeling studies and in vivo pharmacological evaluation of new substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. 2. Effect of the 3-carboxamide substituent on the affinity and selectivity profile. Bioorg Med Chem17:5549-64 (2009) [PubMed] Article Silvestri, R; Ligresti, A; La Regina, G; Piscitelli, F; Gatti, V; Brizzi, A; Pasquini, S; Lavecchia, A; Allarà, M; Fantini, N; Carai, MA; Novellino, E; Colombo, G; Di Marzo, V; Corelli, F Synthesis, cannabinoid receptor affinity, molecular modeling studies and in vivo pharmacological evaluation of new substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. 2. Effect of the 3-carboxamide substituent on the affinity and selectivity profile. Bioorg Med Chem17:5549-64 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cannabinoid receptor 2 | |||
| Name: | Cannabinoid receptor 2 | ||
| Synonyms: | CANNABINOID CB2 | CB-2 | CB2 | CB2A | CB2B | CNR2 | CNR2_HUMAN | CX5 | Cannabinoid CB2 receptor | Cannabinoid receptor 2 (CB2) | Cannabinoid receptor 2 (CB2R) | hCB2 | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 39690.94 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34972 | ||
| Residue: | 360 | ||
| Sequence: |
| ||
| BDBM50298951 | |||
| n/a | |||
| Name | BDBM50298951 | ||
| Synonyms: | CHEMBL577791 | N-[2-(2,4-Dichlorophenyl)ethyl]1-(2,4-dichlorophenyl)-5-(2,5-dimethyl-1H-pyrrol-1-yl)-4-methyl-1H-pyrazole-3-carboxamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C25H22Cl4N4O | ||
| Mol. Mass. | 536.28 | ||
| SMILES | Cc1ccc(C)n1-c1c(C)c(nn1-c1ccc(Cl)cc1Cl)C(=O)NCCc1ccc(Cl)cc1Cl |(32.09,-6.72,;33.01,-5.47,;34.55,-5.47,;35.02,-4,;33.77,-3.09,;33.76,-1.54,;32.52,-4,;31.05,-3.52,;30.58,-2.06,;31.48,-.81,;29.03,-2.06,;28.56,-3.54,;29.8,-4.43,;29.81,-5.98,;31.15,-6.76,;31.15,-8.3,;29.82,-9.07,;29.82,-10.61,;28.48,-8.3,;28.48,-6.76,;27.14,-5.99,;28.12,-.82,;28.74,.59,;26.58,-.98,;25.67,.26,;24.14,.1,;23.23,1.35,;23.86,2.76,;22.94,4,;21.41,3.84,;20.5,5.09,;20.79,2.42,;21.69,1.18,;21.07,-.23,)| | ||
| Structure | 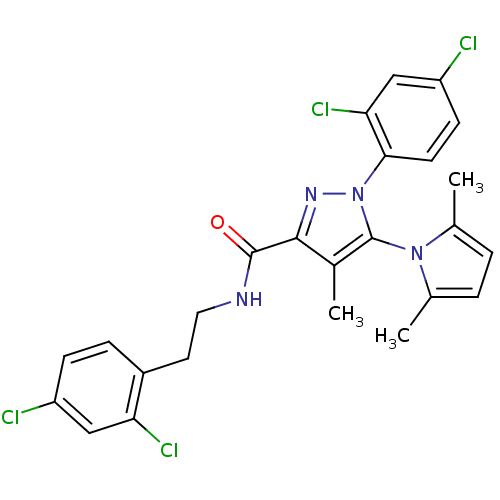
| ||