| Citation |  Silvestri, R; Ligresti, A; La Regina, G; Piscitelli, F; Gatti, V; Brizzi, A; Pasquini, S; Lavecchia, A; Allarà, M; Fantini, N; Carai, MA; Novellino, E; Colombo, G; Di Marzo, V; Corelli, F Synthesis, cannabinoid receptor affinity, molecular modeling studies and in vivo pharmacological evaluation of new substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. 2. Effect of the 3-carboxamide substituent on the affinity and selectivity profile. Bioorg Med Chem17:5549-64 (2009) [PubMed] Article Silvestri, R; Ligresti, A; La Regina, G; Piscitelli, F; Gatti, V; Brizzi, A; Pasquini, S; Lavecchia, A; Allarà, M; Fantini, N; Carai, MA; Novellino, E; Colombo, G; Di Marzo, V; Corelli, F Synthesis, cannabinoid receptor affinity, molecular modeling studies and in vivo pharmacological evaluation of new substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. 2. Effect of the 3-carboxamide substituent on the affinity and selectivity profile. Bioorg Med Chem17:5549-64 (2009) [PubMed] Article |
|---|
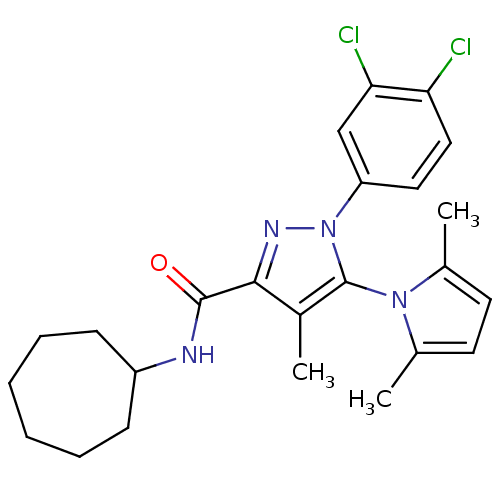
| SMILES | Cc1ccc(C)n1-c1c(C)c(nn1-c1ccc(Cl)c(Cl)c1)C(=O)NC1CCCCCC1 |(31.5,-24.15,;32.38,-22.88,;33.91,-22.86,;34.37,-21.39,;33.1,-20.5,;33.07,-18.96,;31.88,-21.43,;30.4,-20.98,;29.9,-19.53,;30.78,-18.26,;28.36,-19.56,;27.91,-21.02,;29.17,-21.91,;29.2,-23.45,;27.88,-24.24,;27.91,-25.78,;29.26,-26.52,;29.29,-28.06,;30.58,-25.73,;31.92,-26.48,;30.55,-24.2,;27.44,-18.33,;28.03,-16.91,;25.91,-18.52,;24.96,-17.28,;23.48,-17.59,;22.3,-16.6,;22.34,-15.06,;23.56,-14.14,;25.05,-14.51,;25.71,-15.94,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Silvestri, R; Ligresti, A; La Regina, G; Piscitelli, F; Gatti, V; Brizzi, A; Pasquini, S; Lavecchia, A; Allarà, M; Fantini, N; Carai, MA; Novellino, E; Colombo, G; Di Marzo, V; Corelli, F Synthesis, cannabinoid receptor affinity, molecular modeling studies and in vivo pharmacological evaluation of new substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. 2. Effect of the 3-carboxamide substituent on the affinity and selectivity profile. Bioorg Med Chem17:5549-64 (2009) [PubMed] Article
Silvestri, R; Ligresti, A; La Regina, G; Piscitelli, F; Gatti, V; Brizzi, A; Pasquini, S; Lavecchia, A; Allarà, M; Fantini, N; Carai, MA; Novellino, E; Colombo, G; Di Marzo, V; Corelli, F Synthesis, cannabinoid receptor affinity, molecular modeling studies and in vivo pharmacological evaluation of new substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. 2. Effect of the 3-carboxamide substituent on the affinity and selectivity profile. Bioorg Med Chem17:5549-64 (2009) [PubMed] Article