| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Carbonic anhydrase 13 |
|---|
| Ligand | BDBM50303498 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_595959 (CHEMBL1044461) |
|---|
| Ki | 93±n/a nM |
|---|
| Citation |  Maresca, A; Temperini, C; Pochet, L; Masereel, B; Scozzafava, A; Supuran, CT Deciphering the mechanism of carbonic anhydrase inhibition with coumarins and thiocoumarins. J Med Chem53:335-44 (2010) [PubMed] Article Maresca, A; Temperini, C; Pochet, L; Masereel, B; Scozzafava, A; Supuran, CT Deciphering the mechanism of carbonic anhydrase inhibition with coumarins and thiocoumarins. J Med Chem53:335-44 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Carbonic anhydrase 13 |
|---|
| Name: | Carbonic anhydrase 13 |
|---|
| Synonyms: | CA-XIII | CAH13_MOUSE | Ca13 | Car13 | Carbonate dehydratase XIII | Carbonic anhydrase 13 | Carbonic anhydrase XIII |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 29522.80 |
|---|
| Organism: | Mus musculus (mouse) |
|---|
| Description: | Murine cloned isozyme |
|---|
| Residue: | 262 |
|---|
| Sequence: | MARLSWGYGEHNGPIHWNELFPIADGDQQSPIEIKTKEVKYDSSLRPLSIKYDPASAKII
SNSGHSFNVDFDDTEDKSVLRGGPLTGNYRLRQFHLHWGSADDHGSEHVVDGVRYAAELH
VVHWNSDKYPSFVEAAHESDGLAVLGVFLQIGEHNPQLQKITDILDSIKEKGKQTRFTNF
DPLCLLPSSWDYWTYPGSLTVPPLLESVTWIVLKQPISISSQQLARFRSLLCTAEGESAA
FLLSNHRPPQPLKGRRVRASFY
|
|
|
|---|
| BDBM50303498 |
|---|
| n/a |
|---|
| Name | BDBM50303498 |
|---|
| Synonyms: | 1-(2-Oxo-2H-chromen-6-ylmethyl)-3,5,7-triaza-1-azonia-tricyclo[3.3.1.1*3,7*]decane | CHEMBL578167 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H19N4O2 |
|---|
| Mol. Mass. | 299.3471 |
|---|
| SMILES | O=c1ccc2cc(C[N+]34CN5CN(CN(C5)C3)C4)ccc2o1 |TLB:15:14:17:10.9.11,15:10:17:14.16.13,THB:9:8:10.11.15:13,9:10:8.16.17:13| |
|---|
| Structure | 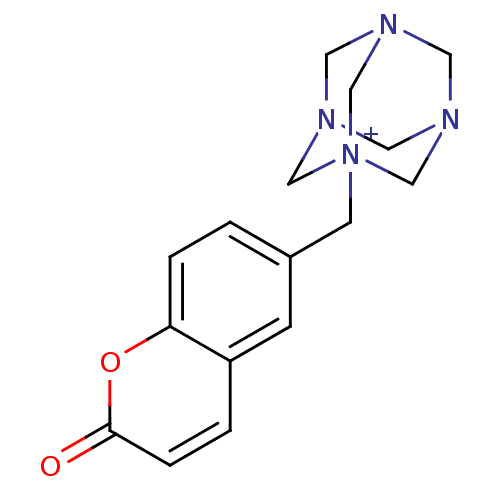
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Maresca, A; Temperini, C; Pochet, L; Masereel, B; Scozzafava, A; Supuran, CT Deciphering the mechanism of carbonic anhydrase inhibition with coumarins and thiocoumarins. J Med Chem53:335-44 (2010) [PubMed] Article
Maresca, A; Temperini, C; Pochet, L; Masereel, B; Scozzafava, A; Supuran, CT Deciphering the mechanism of carbonic anhydrase inhibition with coumarins and thiocoumarins. J Med Chem53:335-44 (2010) [PubMed] Article