| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Uridine 5'-monophosphate synthase |
|---|
| Ligand | BDBM50341906 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_743290 (CHEMBL1767489) |
|---|
| Ki | >4000000±n/a nM |
|---|
| Citation |  Lewis, M; Meza-Avina, ME; Wei, L; Crandall, IE; Bello, AM; Poduch, E; Liu, Y; Paige, CJ; Kain, KC; Pai, EF; Kotra, LP Novel interactions of fluorinated nucleotide derivatives targeting orotidine 5'-monophosphate decarboxylase. J Med Chem54:2891-901 (2011) [PubMed] Article Lewis, M; Meza-Avina, ME; Wei, L; Crandall, IE; Bello, AM; Poduch, E; Liu, Y; Paige, CJ; Kain, KC; Pai, EF; Kotra, LP Novel interactions of fluorinated nucleotide derivatives targeting orotidine 5'-monophosphate decarboxylase. J Med Chem54:2891-901 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Uridine 5'-monophosphate synthase |
|---|
| Name: | Uridine 5'-monophosphate synthase |
|---|
| Synonyms: | Orotate phosphoribosyltransferase (HsOPRT) | Orotidine Monophosphate Decarboxylase (ODCase) | UMPS | UMPS_HUMAN |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52224.99 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P11172 |
|---|
| Residue: | 480 |
|---|
| Sequence: | MAVARAALGPLVTGLYDVQAFKFGDFVLKSGLSSPIYIDLRGIVSRPRLLSQVADILFQT
AQNAGISFDTVCGVPYTALPLATVICSTNQIPMLIRRKETKDYGTKRLVEGTINPGETCL
IIEDVVTSGSSVLETVEVLQKEGLKVTDAIVLLDREQGGKDKLQAHGIRLHSVCTLSKML
EILEQQKKVDAETVGRVKRFIQENVFVAANHNGSPLSIKEAPKELSFGARAELPRIHPVA
SKLLRLMQKKETNLCLSADVSLARELLQLADALGPSICMLKTHVDILNDFTLDVMKELIT
LAKCHEFLIFEDRKFADIGNTVKKQYEGGIFKIASWADLVNAHVVPGSGVVKGLQEVGLP
LHRGCLLIAEMSSTGSLATGDYTRAAVRMAEEHSEFVVGFISGSRVSMKPEFLHLTPGVQ
LEAGGDNLGQQYNSPQEVIGKRGSDIIIVGRGIISAADRLEAAEMYRKAAWEAYLSRLGV
|
|
|
|---|
| BDBM50341906 |
|---|
| n/a |
|---|
| Name | BDBM50341906 |
|---|
| Synonyms: | Ammonium 6-Amido-2'-deoxy-2'-fluoro-beta-D-uridine 5'-O-Monophosphate | CHEMBL1765121 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H11FN3O8P |
|---|
| Mol. Mass. | 351.1829 |
|---|
| SMILES | NC(=O)c1cc(=O)[nH]c(=O)n1[C@@H]1O[C@H](CP([O-])([O-])=O)[C@@H](O)[C@H]1F |r| |
|---|
| Structure | 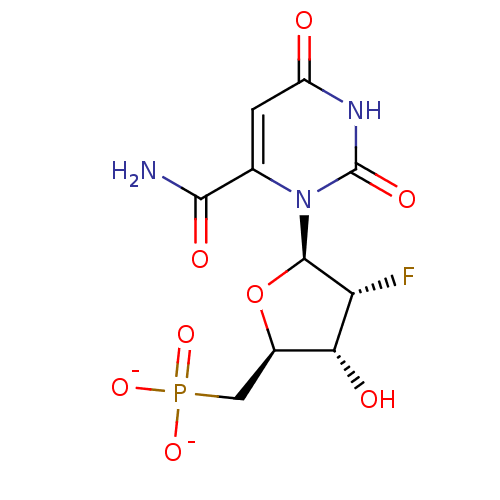
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lewis, M; Meza-Avina, ME; Wei, L; Crandall, IE; Bello, AM; Poduch, E; Liu, Y; Paige, CJ; Kain, KC; Pai, EF; Kotra, LP Novel interactions of fluorinated nucleotide derivatives targeting orotidine 5'-monophosphate decarboxylase. J Med Chem54:2891-901 (2011) [PubMed] Article
Lewis, M; Meza-Avina, ME; Wei, L; Crandall, IE; Bello, AM; Poduch, E; Liu, Y; Paige, CJ; Kain, KC; Pai, EF; Kotra, LP Novel interactions of fluorinated nucleotide derivatives targeting orotidine 5'-monophosphate decarboxylase. J Med Chem54:2891-901 (2011) [PubMed] Article