| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 1 |
|---|
| Ligand | BDBM50350824 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_763305 (CHEMBL1820399) |
|---|
| IC50 | 8±n/a nM |
|---|
| Citation |  Shultz, MD; Cao, X; Chen, CH; Cho, YS; Davis, NR; Eckman, J; Fan, J; Fekete, A; Firestone, B; Flynn, J; Green, J; Growney, JD; Holmqvist, M; Hsu, M; Jansson, D; Jiang, L; Kwon, P; Liu, G; Lombardo, F; Lu, Q; Majumdar, D; Meta, C; Perez, L; Pu, M; Ramsey, T; Remiszewski, S; Skolnik, S; Traebert, M; Urban, L; Uttamsingh, V; Wang, P; Whitebread, S; Whitehead, L; Yan-Neale, Y; Yao, YM; Zhou, L; Atadja, P Optimization of the in vitro cardiac safety of hydroxamate-based histone deacetylase inhibitors. J Med Chem54:4752-72 (2011) [PubMed] Article Shultz, MD; Cao, X; Chen, CH; Cho, YS; Davis, NR; Eckman, J; Fan, J; Fekete, A; Firestone, B; Flynn, J; Green, J; Growney, JD; Holmqvist, M; Hsu, M; Jansson, D; Jiang, L; Kwon, P; Liu, G; Lombardo, F; Lu, Q; Majumdar, D; Meta, C; Perez, L; Pu, M; Ramsey, T; Remiszewski, S; Skolnik, S; Traebert, M; Urban, L; Uttamsingh, V; Wang, P; Whitebread, S; Whitehead, L; Yan-Neale, Y; Yao, YM; Zhou, L; Atadja, P Optimization of the in vitro cardiac safety of hydroxamate-based histone deacetylase inhibitors. J Med Chem54:4752-72 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 1 |
|---|
| Name: | Histone deacetylase 1 |
|---|
| Synonyms: | Cereblon/Histone deacetylase 1 | HD1 | HDAC1 | HDAC1_HUMAN | Histone deacetylase 1 (HDAC1) | Human HDAC1 | RPD3L1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55090.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13547 |
|---|
| Residue: | 482 |
|---|
| Sequence: | MAQTQGTRRKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKAN
AEEMTKYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGEDCPVFDGLFEFCQLSTGGSVAS
AVKLNKQQTDIAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYIDIDIHHG
DGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGIDDESYEAI
FKPVMSKVMEMFQPSAVVLQCGSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNLPMLMLGG
GGYTIRNVARCWTYETAVALDTEIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTNEYLE
KIKQRLFENLRMLPHAPGVQMQAIPEDAIPEESGDEDEDDPDKRISICSSDKRIACEEEF
SDSEEEGEGGRKNSSNFKKAKRVKTEDEKEKDPEEKKEVTEEEKTKEEKPEAKGVKEEVK
LA
|
|
|
|---|
| BDBM50350824 |
|---|
| n/a |
|---|
| Name | BDBM50350824 |
|---|
| Synonyms: | CHEMBL1819264 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H21N5O2 |
|---|
| Mol. Mass. | 351.4023 |
|---|
| SMILES | Cc1[nH]c2nccnc2c1CCNCc1ccc(C=CC(=O)NO)cc1 |w:18.19| |
|---|
| Structure | 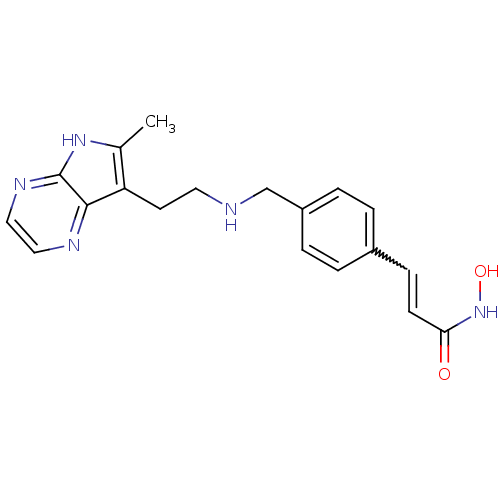
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Shultz, MD; Cao, X; Chen, CH; Cho, YS; Davis, NR; Eckman, J; Fan, J; Fekete, A; Firestone, B; Flynn, J; Green, J; Growney, JD; Holmqvist, M; Hsu, M; Jansson, D; Jiang, L; Kwon, P; Liu, G; Lombardo, F; Lu, Q; Majumdar, D; Meta, C; Perez, L; Pu, M; Ramsey, T; Remiszewski, S; Skolnik, S; Traebert, M; Urban, L; Uttamsingh, V; Wang, P; Whitebread, S; Whitehead, L; Yan-Neale, Y; Yao, YM; Zhou, L; Atadja, P Optimization of the in vitro cardiac safety of hydroxamate-based histone deacetylase inhibitors. J Med Chem54:4752-72 (2011) [PubMed] Article
Shultz, MD; Cao, X; Chen, CH; Cho, YS; Davis, NR; Eckman, J; Fan, J; Fekete, A; Firestone, B; Flynn, J; Green, J; Growney, JD; Holmqvist, M; Hsu, M; Jansson, D; Jiang, L; Kwon, P; Liu, G; Lombardo, F; Lu, Q; Majumdar, D; Meta, C; Perez, L; Pu, M; Ramsey, T; Remiszewski, S; Skolnik, S; Traebert, M; Urban, L; Uttamsingh, V; Wang, P; Whitebread, S; Whitehead, L; Yan-Neale, Y; Yao, YM; Zhou, L; Atadja, P Optimization of the in vitro cardiac safety of hydroxamate-based histone deacetylase inhibitors. J Med Chem54:4752-72 (2011) [PubMed] Article