| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Ligand | BDBM24466 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_819215 (CHEMBL2032484) |
|---|
| IC50 | 100000±n/a nM |
|---|
| Citation |  Liu, L; Norman, MH; Lee, M; Xi, N; Siegmund, A; Boezio, AA; Booker, S; Choquette, D; D'Angelo, ND; Germain, J; Yang, K; Yang, Y; Zhang, Y; Bellon, SF; Whittington, DA; Harmange, JC; Dominguez, C; Kim, TS; Dussault, I Structure-based design of novel class II c-Met inhibitors: 2. SAR and kinase selectivity profiles of the pyrazolone series. J Med Chem55:1868-97 (2012) [PubMed] Article Liu, L; Norman, MH; Lee, M; Xi, N; Siegmund, A; Boezio, AA; Booker, S; Choquette, D; D'Angelo, ND; Germain, J; Yang, K; Yang, Y; Zhang, Y; Bellon, SF; Whittington, DA; Harmange, JC; Dominguez, C; Kim, TS; Dussault, I Structure-based design of novel class II c-Met inhibitors: 2. SAR and kinase selectivity profiles of the pyrazolone series. J Med Chem55:1868-97 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Name: | cAMP-dependent protein kinase catalytic subunit alpha |
|---|
| Synonyms: | KAPCA_HUMAN | PKA C-alpha | PKACA | PRKACA | cAMP-dependent protein kinase (PKA) | cAMP-dependent protein kinase catalytic (PKA) | cAMP-dependent protein kinase catalytic subunit alpha | cAMP-dependent protein kinase catalytic subunit alpha (PKA) | cAMP-dependent protein kinase catalytic subunit alpha (PKACA) | cAMP-dependent protein kinase catalytic subunit alpha (PKAc) | cAMP-dependent protein kinase catalytic subunit alpha isoform 1 | cAMP-dependent protein kinase, alpha-catalytic subunit |
|---|
| Type: | Enzyme Catalytic Subunit |
|---|
| Mol. Mass.: | 40598.73 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 351 |
|---|
| Sequence: | MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVML
VKHKETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV
MEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY
IQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF
ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT
TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF
|
|
|
|---|
| BDBM24466 |
|---|
| n/a |
|---|
| Name | BDBM24466 |
|---|
| Synonyms: | 1-(2-hydroxy-2-methylpropyl)-N-{5-[(7-methoxyquinolin-4-yl)oxy]pyridin-2-yl}-5-methyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | Substituted Pyrazolone, 17 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C30H29N5O5 |
|---|
| Mol. Mass. | 539.5818 |
|---|
| SMILES | COc1ccc2c(Oc3ccc(NC(=O)c4c(C)n(CC(C)(C)O)n(-c5ccccc5)c4=O)nc3)ccnc2c1 |
|---|
| Structure | 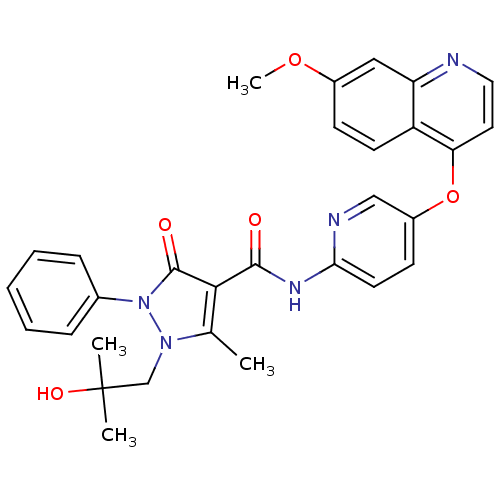
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Liu, L; Norman, MH; Lee, M; Xi, N; Siegmund, A; Boezio, AA; Booker, S; Choquette, D; D'Angelo, ND; Germain, J; Yang, K; Yang, Y; Zhang, Y; Bellon, SF; Whittington, DA; Harmange, JC; Dominguez, C; Kim, TS; Dussault, I Structure-based design of novel class II c-Met inhibitors: 2. SAR and kinase selectivity profiles of the pyrazolone series. J Med Chem55:1868-97 (2012) [PubMed] Article
Liu, L; Norman, MH; Lee, M; Xi, N; Siegmund, A; Boezio, AA; Booker, S; Choquette, D; D'Angelo, ND; Germain, J; Yang, K; Yang, Y; Zhang, Y; Bellon, SF; Whittington, DA; Harmange, JC; Dominguez, C; Kim, TS; Dussault, I Structure-based design of novel class II c-Met inhibitors: 2. SAR and kinase selectivity profiles of the pyrazolone series. J Med Chem55:1868-97 (2012) [PubMed] Article