| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Ephrin type-B receptor 1 |
|---|
| Ligand | BDBM50389320 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_831783 (CHEMBL2065146) |
|---|
| IC50 | <10000±n/a nM |
|---|
| Citation |  Maryanoff, BE; O'Neill, JC; McComsey, DF; Yabut, SC; Luci, DK; Gibbs, AC; Connelly, MA Pyrimidinopyrimidine inhibitors of ketohexokinase: exploring the ring C2 group that interacts with Asp-27B in the ligand binding pocket. Bioorg Med Chem Lett22:5326-9 (2012) [PubMed] Article Maryanoff, BE; O'Neill, JC; McComsey, DF; Yabut, SC; Luci, DK; Gibbs, AC; Connelly, MA Pyrimidinopyrimidine inhibitors of ketohexokinase: exploring the ring C2 group that interacts with Asp-27B in the ligand binding pocket. Bioorg Med Chem Lett22:5326-9 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Ephrin type-B receptor 1 |
|---|
| Name: | Ephrin type-B receptor 1 |
|---|
| Synonyms: | ELK | EPH receptor B1 | EPHB1 | EPHB1_HUMAN | EPHT2 | Ephrin receptor | Ephrin type-B receptor 1 | HEK6 | NET |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 109882.53 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | EPH receptor B1 EPHB1 HUMAN::P54762 |
|---|
| Residue: | 984 |
|---|
| Sequence: | MALDYLLLLLLASAVAAMEETLMDTRTATAELGWTANPASGWEEVSGYDENLNTIRTYQV
CNVFEPNQNNWLLTTFINRRGAHRIYTEMRFTVRDCSSLPNVPGSCKETFNLYYYETDSV
IATKKSAFWSEAPYLKVDTIAADESFSQVDFGGRLMKVNTEVRSFGPLTRNGFYLAFQDY
GACMSLLSVRVFFKKCPSIVQNFAVFPETMTGAESTSLVIARGTCIPNAEEVDVPIKLYC
NGDGEWMVPIGRCTCKPGYEPENSVACKACPAGTFKASQEAEGCSHCPSNSRSPAEASPI
CTCRTGYYRADFDPPEVACTSVPSGPRNVISIVNETSIILEWHPPRETGGRDDVTYNIIC
KKCRADRRSCSRCDDNVEFVPRQLGLTECRVSISSLWAHTPYTFDIQAINGVSSKSPFPP
QHVSVNITTNQAAPSTVPIMHQVSATMRSITLSWPQPEQPNGIILDYEIRYYEKEHNEFN
SSMARSQTNTARIDGLRPGMVYVVQVRARTVAGYGKFSGKMCFQTLTDDDYKSELREQLP
LIAGSAAAGVVFVVSLVAISIVCSRKRAYSKEAVYSDKLQHYSTGRGSPGMKIYIDPFTY
EDPNEAVREFAKEIDVSFVKIEEVIGAGEFGEVYKGRLKLPGKREIYVAIKTLKAGYSEK
QRRDFLSEASIMGQFDHPNIIRLEGVVTKSRPVMIITEFMENGALDSFLRQNDGQFTVIQ
LVGMLRGIAAGMKYLAEMNYVHRDLAARNILVNSNLVCKVSDFGLSRYLQDDTSDPTYTS
SLGGKIPVRWTAPEAIAYRKFTSASDVWSYGIVMWEVMSFGERPYWDMSNQDVINAIEQD
YRLPPPMDCPAALHQLMLDCWQKDRNSRPRFAEIVNTLDKMIRNPASLKTVATITAVPSQ
PLLDRSIPDFTAFTTVDDWLSAIKMVQYRDSFLTAGFTSLQLVTQMTSEDLLRIGITLAG
HQKKILNSIHSMRVQISQSPTAMA
|
|
|
|---|
| BDBM50389320 |
|---|
| n/a |
|---|
| Name | BDBM50389320 |
|---|
| Synonyms: | CHEMBL2063926 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H35N9S |
|---|
| Mol. Mass. | 505.681 |
|---|
| SMILES | CSc1ccccc1Nc1nc(nc2c(NCC3CC3)ncnc12)N1CCN(CC1)C1CCNCC1 |
|---|
| Structure | 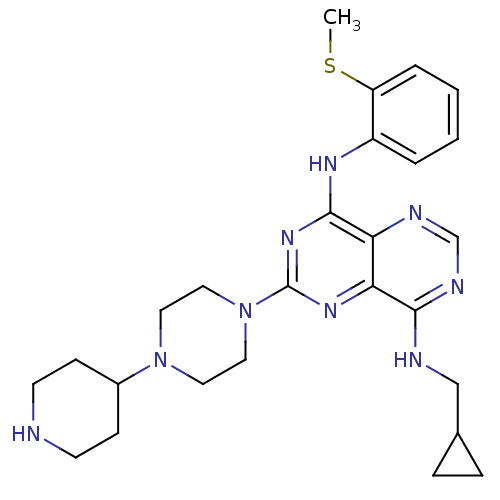
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Maryanoff, BE; O'Neill, JC; McComsey, DF; Yabut, SC; Luci, DK; Gibbs, AC; Connelly, MA Pyrimidinopyrimidine inhibitors of ketohexokinase: exploring the ring C2 group that interacts with Asp-27B in the ligand binding pocket. Bioorg Med Chem Lett22:5326-9 (2012) [PubMed] Article
Maryanoff, BE; O'Neill, JC; McComsey, DF; Yabut, SC; Luci, DK; Gibbs, AC; Connelly, MA Pyrimidinopyrimidine inhibitors of ketohexokinase: exploring the ring C2 group that interacts with Asp-27B in the ligand binding pocket. Bioorg Med Chem Lett22:5326-9 (2012) [PubMed] Article