| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dipeptidyl peptidase 9 |
|---|
| Ligand | BDBM50389475 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_832307 (CHEMBL2066845) |
|---|
| IC50 | 9.3±n/a nM |
|---|
| Citation |  Wu, W; Liu, Y; Milo, LJ; Shu, Y; Zhao, P; Li, Y; Woznica, I; Yu, G; Sanford, DG; Zhou, Y; Poplawski, SE; Connolly, BA; Sudmeier, JL; Bachovchin, WW; Lai, JH 4-Substituted boro-proline dipeptides: synthesis, characterization, and dipeptidyl peptidase IV, 8, and 9 activities. Bioorg Med Chem Lett22:5536-40 (2012) [PubMed] Article Wu, W; Liu, Y; Milo, LJ; Shu, Y; Zhao, P; Li, Y; Woznica, I; Yu, G; Sanford, DG; Zhou, Y; Poplawski, SE; Connolly, BA; Sudmeier, JL; Bachovchin, WW; Lai, JH 4-Substituted boro-proline dipeptides: synthesis, characterization, and dipeptidyl peptidase IV, 8, and 9 activities. Bioorg Med Chem Lett22:5536-40 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dipeptidyl peptidase 9 |
|---|
| Name: | Dipeptidyl peptidase 9 |
|---|
| Synonyms: | DPP9 | DPP9_HUMAN | DPRP-2 | DPRP2 | Dipeptidyl peptidase 9 (DDP9) | Dipeptidyl peptidase 9 (DPP-9) | Dipeptidyl peptidase 9 (DPP9) | Dipeptidyl peptidase IV-related protein 2 | Dipeptidyl peptidase IX | Dipeptidyl peptidase IX (DDP-IX) | Dipeptidyl peptidase-like protein 9 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 98260.70 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q86TI2 |
|---|
| Residue: | 863 |
|---|
| Sequence: | MATTGTPTADRGDAAATDDPAARFQVQKHSWDGLRSIIHGSRKYSGLIVNKAPHDFQFVQ
KTDESGPHSHRLYYLGMPYGSRENSLLYSEIPKKVRKEALLLLSWKQMLDHFQATPHHGV
YSREEELLRERKRLGVFGITSYDFHSESGLFLFQASNSLFHCRDGGKNGFMVSPMKPLEI
KTQCSGPRMDPKICPADPAFFSFINNSDLWVANIETGEERRLTFCHQGLSNVLDDPKSAG
VATFVIQEEFDRFTGYWWCPTASWEGSEGLKTLRILYEEVDESEVEVIHVPSPALEERKT
DSYRYPRTGSKNPKIALKLAEFQTDSQGKIVSTQEKELVQPFSSLFPKVEYIARAGWTRD
GKYAWAMFLDRPQQWLQLVLLPPALFIPSTENEEQRLASARAVPRNVQPYVVYEEVTNVW
INVHDIFYPFPQSEGEDELCFLRANECKTGFCHLYKVTAVLKSQGYDWSEPFSPGEDEFK
CPIKEEIALTSGEWEVLARHGSKIWVNEETKLVYFQGTKDTPLEHHLYVVSYEAAGEIVR
LTTPGFSHSCSMSQNFDMFVSHYSSVSTPPCVHVYKLSGPDDDPLHKQPRFWASMMEAAS
CPPDYVPPEIFHFHTRSDVRLYGMIYKPHALQPGKKHPTVLFVYGGPQVQLVNNSFKGIK
YLRLNTLASLGYAVVVIDGRGSCQRGLRFEGALKNQMGQVEIEDQVEGLQFVAEKYGFID
LSRVAIHGWSYGGFLSLMGLIHKPQVFKVAIAGAPVTVWMAYDTGYTERYMDVPENNQHG
YEAGSVALHVEKLPNEPNRLLILHGFLDENVHFFHTNFLVSQLIRAGKPYQLQIYPNERH
SIRCPESGEHYEVTLLHFLQEYL
|
|
|
|---|
| BDBM50389475 |
|---|
| n/a |
|---|
| Name | BDBM50389475 |
|---|
| Synonyms: | CHEMBL2063035 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C9H17BN2O5 |
|---|
| Mol. Mass. | 244.053 |
|---|
| SMILES | OB(O)[C@@H]1C[C@H](O)CN1C(=O)[C@@H]1C[C@@H](O)CN1 |r| |
|---|
| Structure | 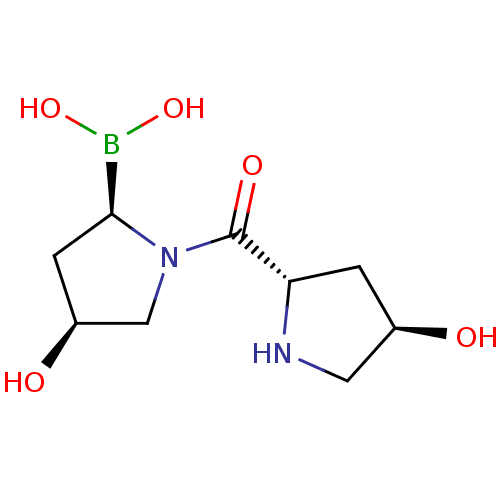
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wu, W; Liu, Y; Milo, LJ; Shu, Y; Zhao, P; Li, Y; Woznica, I; Yu, G; Sanford, DG; Zhou, Y; Poplawski, SE; Connolly, BA; Sudmeier, JL; Bachovchin, WW; Lai, JH 4-Substituted boro-proline dipeptides: synthesis, characterization, and dipeptidyl peptidase IV, 8, and 9 activities. Bioorg Med Chem Lett22:5536-40 (2012) [PubMed] Article
Wu, W; Liu, Y; Milo, LJ; Shu, Y; Zhao, P; Li, Y; Woznica, I; Yu, G; Sanford, DG; Zhou, Y; Poplawski, SE; Connolly, BA; Sudmeier, JL; Bachovchin, WW; Lai, JH 4-Substituted boro-proline dipeptides: synthesis, characterization, and dipeptidyl peptidase IV, 8, and 9 activities. Bioorg Med Chem Lett22:5536-40 (2012) [PubMed] Article